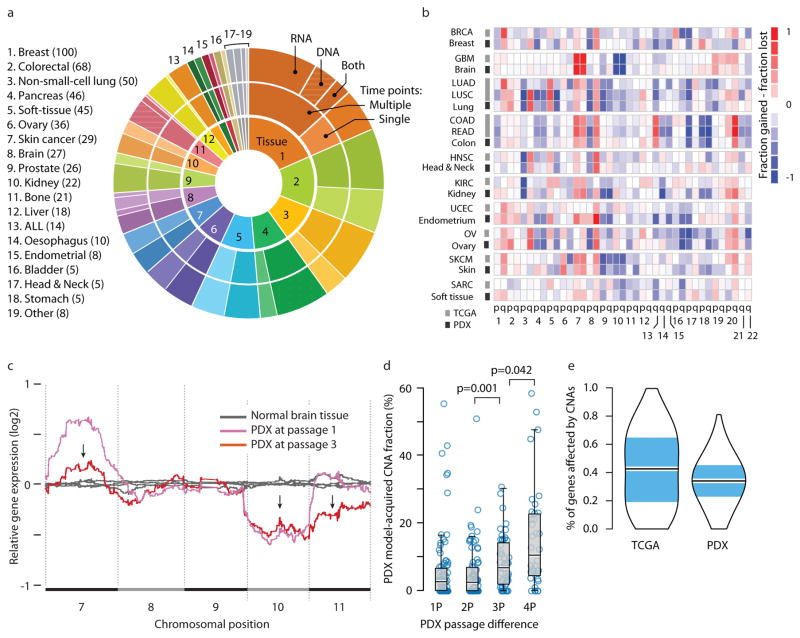
Figure 1. The landscape of aneuploidy and copy number alterations in PDXs.
(a) Distribution of cancer types in our PDX dataset (n=543 unique models). In the inner circle models are divided by their lineage: each cancer type is denoted by a color and a number. In the middle circle models are divided by the number of time points analyzed: multiple time points are denoted by a darker color, and enable to follow PDX evolution throughout in vivo propagation. In the outer circle models are divided by the biological material from which CNAs were inferred: DNA (stripes), RNA (dots) or both (stripes and dots). (b) A heatmap comparing the landscapes of lineage-matched arm-level CNAs of PDXs and of primary TCGA tumors, showing an overall high degree of concordance (mean Pearson’s r = 0.79). The color of each chromosome arm represents the fraction difference between gains and losses of that arm. (c) A representative example of PDX model evolution. Shown are gene expression moving average plots of normal brain tissue (gray), GBM PDX model at p1 (pink) and GBM model at p3 (red), revealing the disappearance of trisomy 7, the retention of monosomy 10, and the emergence of monosomy 11, within two in vivo passages. (d) Gradual evolution of CNA landscapes throughout PDX passaging. Box plots present model-acquired CNA fraction as a function of the number of passages between measurements. Bar, median; box, 25th and 75th percentiles; whiskers, data within 1.5*IQR of lower or upper quartile; circles: all data points. P-values indicate significance from a Wilcoxon rank-sum test. (e) Violin plots present the proportion of genes affected by CNAs in TCGA and in PDX tumor samples (all tissue types combined), showing an overall similarity between both datasets. Bar, median; colored rectangle, 25th and 75th percentiles; width of the violin indicates frequency at that CNA fraction level.