ABSTRACT
Newly assembled herpesvirus nucleocapsids are translocated from the nucleus to the cytosol by a vesicle-mediated process engaging the nuclear membranes. This transport is governed by the conserved nuclear egress complex (NEC), consisting of the alphaherpesviral pUL34 and pUL31 homologs. The NEC is not only required for efficient nuclear egress but also sufficient for vesicle formation from the inner nuclear membrane (INM), as well as from synthetic lipid bilayers. The recently solved crystal structures for the NECs from different herpesviruses revealed molecular details of this membrane deformation and scission machinery uncovering the interfaces involved in complex and coat formation. However, the interaction domain with the nucleocapsid remained undefined. Since the NEC assembles a curved hexagonal coat on the nucleoplasmic side of the INM consisting of tightly interwoven pUL31/pUL34 heterodimers arranged in hexamers, only the membrane-distal end of the NEC formed by pUL31 residues appears to be accessible for interaction with the nucleocapsid cargo. To identify the amino acids involved in capsid incorporation, we mutated the corresponding regions in the alphaherpesvirus pseudorabies virus (PrV). Site-specifically mutated pUL31 homologs were tested for localization, interaction with pUL34, and complementation of PrV-ΔUL31. We identified a conserved lysine residue at amino acid position 242 in PrV pUL31 located in the alpha-helical domain H10 exposed on the membrane-distal end of the NEC as a key residue for nucleocapsid incorporation into the nascent primary particle.
IMPORTANCE Vesicular transport through the nuclear envelope is a focus of research but is still not well understood. Herpesviruses pioneered this mechanism for translocation of the newly assembled nucleocapsid from the nucleus into the cytosol via vesicles derived from the inner nuclear membrane which fuse in a well-tuned process with the outer nuclear membrane to release their content. The structure of the viral nuclear membrane budding and scission machinery has been solved recently, providing in-depth molecular details. However, how cargo is incorporated remained unclear. We identified a conserved lysine residue in the membrane-distal portion of the nuclear egress complex required for capsid uptake into inner nuclear membrane-derived vesicles.
KEYWORDS: herpesvirus, pseudorabies virus (PrV), nuclear egress complex (NEC), pUL31, pUL34, nucleocapsid, NEC capsid interaction
INTRODUCTION
Herpesviruses are large enveloped DNA viruses which use two different subcellular compartments for virion assembly. Whereas capsid formation and genome packaging take place in the nucleus, final virus maturation proceeds in the cytoplasm (1). Although nucleocytoplasmic transport normally is restricted to nuclear pores, size constraints prevent the approximately 125-nm-diameter herpesvirus nucleocapsid from using this pathway. Thus, to overcome the nuclear envelope barrier herpesvirus nucleocapsids engage a vesicular transport process which involves both nuclear membranes. Mature intranuclear capsids bud at the inner nuclear membrane (INM), resulting in a primary enveloped nucleocapsid located in the perinuclear space (PNS). To leave the PNS, the INM-derived primary envelope fuses in a mechanistically as-yet-unknown process with the outer nuclear membrane (ONM), releasing the nucleocapsid into the cytosol (reviewed in references 2, 3, and 4).
The formation of primary virions in the PNS is mediated by the nuclear egress complex (NEC), which is conserved within the Herpesviridae. The major components of the NEC are homologs of the viral proteins designated pUL34 and pUL31 in the alphaherpesviruses pseudorabies virus (PrV) and herpes simplex virus (HSV). The pUL34 homologs are tail-anchored membrane proteins which are efficiently targeted to the nuclear envelope even in the absence of other viral proteins. In contrast, in the absence of pUL34, the pUL31 homologs are diffusely distributed throughout the nucleus. In the presence of and by direct interaction with pUL34, pUL31 is recruited to the INM to form the NEC at the nucleoplasmic face of the nuclear envelope (reviewed in references 2, 3, and 4). The NEC itself is sufficient to mediate membrane bending and vesicle scission not only from the INM in eukaryotic cells but also from synthetic lipid membranes, e.g., giant unilamellar vesicles indicating that no other viral or cellular protein is required for the budding process (5, 6). Interestingly, during herpesvirus infection formation of empty primary envelopes is rarely observed, whereas DNA-filled C capsids constitute the predominant substrate for primary envelopment indicating that NEC oligomerization is precluded until a mature capsid initiates budding (7, 8).
The capsid-associated proteins pUL17 and pUL25, which form the capsid vertex-specific component (CVSC), are enriched on mature capsids and are likely to provide the trigger on C capsids to initiate nuclear export (9, 10). In line with this, a PrV mutant lacking pUL25 forms mature capsids which accumulate in the nucleus in close apposition to the INM but are unable to bud (11). The role of the pUL17 component of the CVSC during nuclear egress is difficult to assess since it is also essential for the mechanistically coupled DNA-encapsidation process and its absence results in the formation of only immature B capsids which contain scaffold but lack viral genome and which are only inefficiently exported from the nucleus (12, 13).
Recent data indicate that pUL31 binds to the capsid already in the nucleoplasm and mediates its translocation to the future budding site (14). However, how interaction between nucleocapsid and pUL31 and/or the NEC occurs remains unclear. A direct interaction between the CVSC and pUL31 was shown for HSV-1 (15, 16), while for PrV an interaction with the nucleocapsid in the absence of the CVSC component pUL25 has been reported (17) pointing to multiple binding partners and sites on the capsid. Due to the transient nature of the nucleocapsid-NEC interaction, which is required for capsid incorporation but has to disintegrate for cargo delivery into the cytoplasm, an unambiguous identification of the interaction partners by biochemical approaches has proven to be difficult.
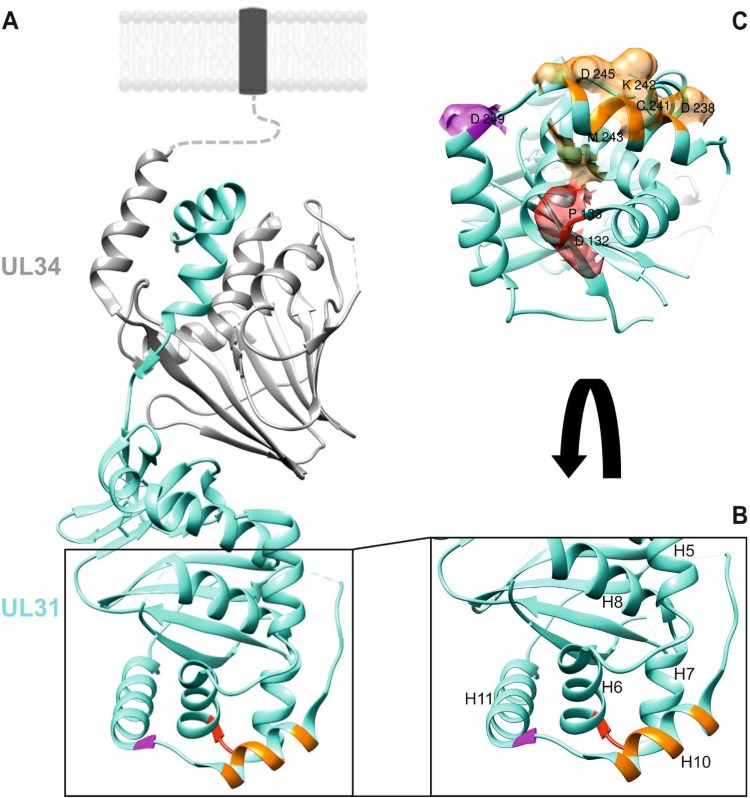
The recently solved structures of the NECs from the alphaherpesviruses HSV-1 and PrV (18, 19) and from the betaherpesvirus human cytomegalovirus (20, 21), as well as a high-resolution imaging approach of cells simultaneously expressing PrV pUL31 and pUL34 (22), revealed molecular details of this viral membrane remodeling and scission machinery. Both proteins possess a general globular fold sitting on top of each other forming an extended rod (see Fig. 1). The N-terminal arm of pUL31 extends around the core of the membrane anchored pUL34 which forms a pedestal at the nucleoplasmic side of the INM. Vesicle formation from the membrane is driven by assembly of the NEC heterodimers into an honeycomb-like coat as shown in vitro (5), as well as in situ in cells simultaneously expressing PrV pUL31 and pUL34 (22). Based on the crystal structures, only the membrane-distal end of the NEC, which is formed by the pUL31 homologs, appears accessible for the nucleocapsid cargo. In addition, it was speculated that cargo binding most likely involves charged residues (19). For PrV pUL31 we assumed that the putative capsid interaction interface might be formed either by a region comprising amino acids (aa) 132 to 134 forming a loop with alpha-helices H6 and H7, including an aspartate (D) at position 132, and/or aa 236 to 245 building a long alpha-helical region (H10), with charged D238, K242, and D245, and/or a loop region (aa 246 to 248) leading into H11 with D249 (see Fig. 1) (19). To assess their importance in capsid incorporation into the nascent primary vesicle, we site-specifically mutated the specific codons and tested the modified pUL31 for location and NEC formation after transient expression, for complementation of PrV-ΔUL31 and for functional nuclear egress of capsids or vesicle formation from the INM after establishing cell lines stably expressing the altered NEC. By this approach, we identified a lysine residue in H10 (K242), which is highly conserved in pUL31 homologs of the alpha- and betaherpesviruses, as a key residue for nucleocapsid incorporation into the INM-derived vesicles.
FIG 1.
Location of putative capsid interaction interfaces in PrV pUL31. (A) In the PrV NEC structure (19), the pUL34 component is shown in gray, and pUL31 is shown in turquoise. Orientation toward and anchorage in the inner nuclear membrane or the primary virion envelope is indicated by the dotted line, and the transmembrane anchor is represented by a dark box. Location of amino acids mutated in this study is indicated in red (D132 and P133), orange (D238, C241, K242, M243, and D245), and magenta (D249). (B) Enlarged membrane-distal end of the NEC in ribbon presentation with alpha-helices numbered, as described previously (19). (C) Amino acids changed in this study are shown in surface presentation using the same coloring. Molecular graphics and analyses were performed with the UCSF Chimera package (35).
RESULTS
Mutational analysis of PrV pUL31.
To test for the capsid binding interface on the NEC, we site-specifically mutated regions which were supposed to be exposed at the membrane-distal end of the NEC. Since it was predicted that capsid interaction is mediated by electrostatic interactions (19), we primarily targeted charged residues. To this end, we singly changed codons for asparagine (D) at position 132 and the neighboring noncharged proline (P) 133, predicted to be part of a small loop between alpha-helix 6 (H6) and H7, D238 and D245 located at the boundaries of H10, as well as D249 in H11 to alanine codons (A). In addition, we mutated the central region of H10 and simultaneously substituted codons for cysteine (C) 241, lysine (K) 242, and methionine (M) 243 for alanine codons (Fig. 1). Since it turned out that the triple mutant showed the expected phenotype, we also substituted only K242 by alanine. Correct mutagenesis was verified by sequencing (data not shown), and the expression of the mutated proteins was demonstrated by indirect immunofluorescence and Western blotting.
NEC formation in transient-transfection assays.
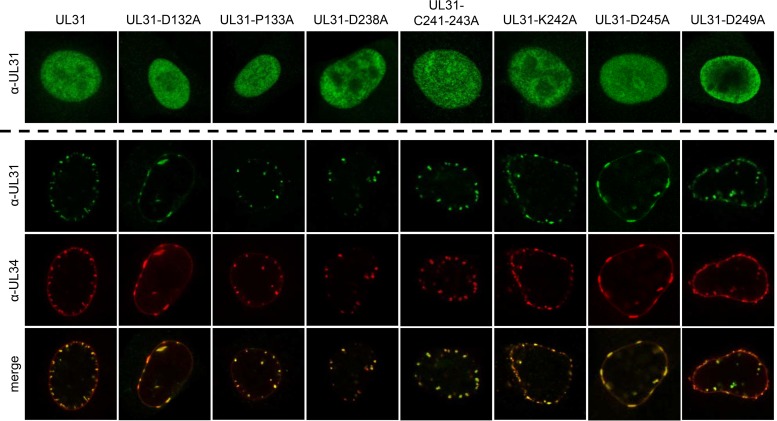
Correct expression and nuclear localization of the mutated pUL31 was tested after transfection of the corresponding expression plasmids into rabbit kidney (RK13) cells (Fig. 2, upper panels). Indirect immunofluorescence with the monospecific anti-UL31 serum showed nuclear localization for all mutants comparable to wild-type pUL31, indicating that overall protein folding, stability, or trafficking was not compromised by the amino acid changes. Since we aimed at mutants that were only deficient in capsid interaction but not in NEC formation or vesicle scission, we analyzed whether the mutated proteins were still able to interact with pUL34 in transient-transfection assays. NEC formation is easily detectable by relocalization of nucleoplasmic pUL31 to the nuclear rim in the presence of pUL34, resulting in typical pUL31/pUL34 costaining punctae in indirect immunofluorescence assays (23, 24). To this end, pcDNA-UL34 was cotransfected with wild-type and mutated pUL31 expression plasmids (Fig. 2, lower panels). After 2 days, the cells were fixed and permeabilized and then simultaneously stained with the monospecific pUL31 antiserum and a pUL34-specific monoclonal antibody. Coexpression of pUL34 resulted in the redistribution of pUL31 from a diffuse nuclear staining (Fig. 2, upper panels) into speckles at or close to the nuclear rim (Fig. 2, lower panels), demonstrating that the pUL31 mutants are still able to interact with the complex partner pUL34. Although pUL31-P133A, pUL31-D238A, pUL31-C241-243A, pUL31-K242A, and pUL31-D249A showed the typical roundish speckle pattern, cotransfection of pcDNA-UL31-D132A and pcDNA-UL31-D245A resulted in more flat, elongated fluorescent structures at the nuclear envelope, indicating that membrane curvature might be affected by the mutations.
FIG 2.
Intracellular localization of mutated pUL31 and colocalization with pUL34. Intracellular localization of the mutated proteins was tested after transfection of RK13 cells with the corresponding pUL31 expression plasmids (upper panels). pUL31 was detected with the monospecific anti-pUL31 serum. Speckle formation indicative of vesicle formation from the inner nuclear membrane was analyzed after cotransfection of the indicated pUL31 expression constructs with pcDNA-UL34. pUL31 was detected by the monospecific anti-pUL31 serum (green), and pUL34 was stained with a monoclonal pUL34-specific antibody (red). Merged images are shown in the lowermost panel. Immunofluorescence was recorded with a confocal laser scanning microscope (SP5; Leica, Germany).
Complementation of PrV-ΔUL31.
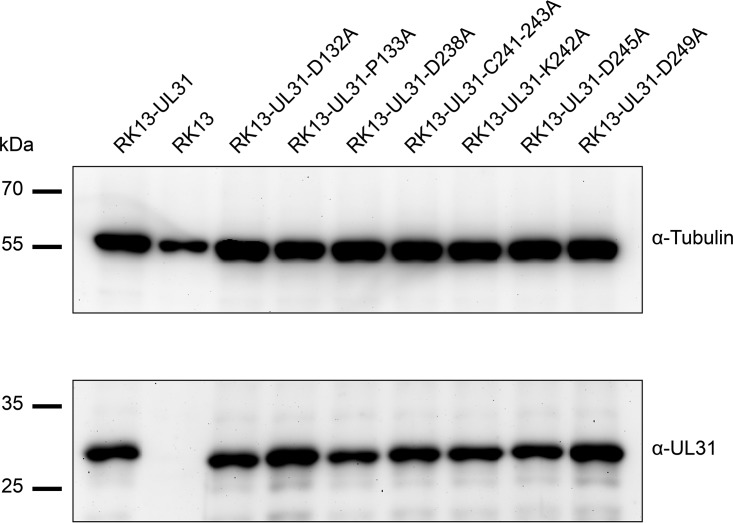
To investigate whether the mutated pUL31 are still able to form a functional NEC and to efficiently translocate capsids from the nucleus to the cytoplasm where maturation to infectious particles can proceed, cell lines were generated which stably express the respective pUL31 mutants. To this end, RK13 cells were transfected with the corresponding pUL31 expression plasmids and selected with G418. Cell colonies were screened for presence of pUL31 by indirect immunofluorescence using the monospecific anti-pUL31 serum (data not shown) and correct protein expression was analyzed by Western blotting (Fig. 3). In contrast to nontransgenic parental RK13 cells, pUL31 was detectable in all selected cell clones expressing native or mutated pUL31.
FIG 3.
pUL31 expression in RK13 cell lines. RK13 cells stably expressing native pUL31 or the mutated proteins, as well as nontransgenic RK13 cells, were harvested by scraping them into the media. Cells were lysed in sample buffer, and proteins were separated in an SDS-10% polyacrylamide gel. After transfer to nitrocellulose, the blot was incubated with the monospecific anti-pUL31 rabbit serum and a monoclonal alpha-tubulin mouse antibody as loading control. Molecular masses of marker proteins are indicated on the left in kilodaltons.
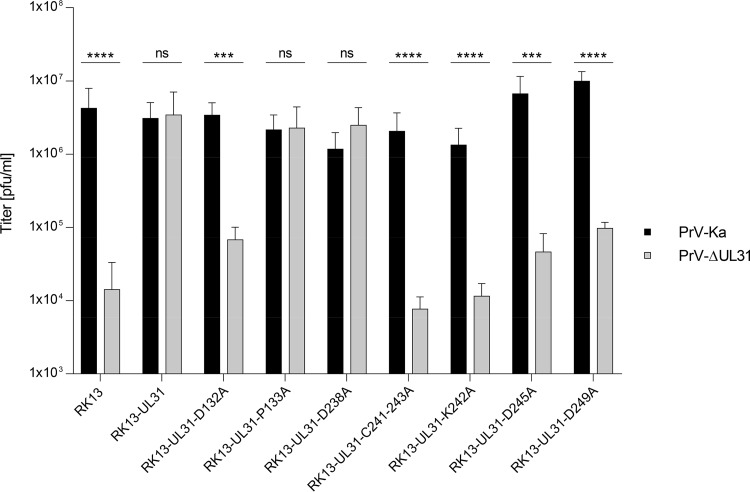
For transcomplementation assays (Fig. 4), RK13 and cell lines expressing pUL31 were infected with PrV strain Kaplan (PrV-Ka) or the UL31 deletion mutant PrV-ΔUL31 at a multiplicity of infection (MOI) of 3, and cells and supernatants were harvested 24 h postinfection (p.i.). Progeny virus titers were determined on transcomplementing RK13-UL31 cells. Infection with PrV-Ka resulted in titers between 106 and 107 PFU per ml (PFU/ml) from all cell lines tested indicating that none of the expressed pUL31 mutants exerted a dominant-negative effect. After infection with PrV-ΔUL31, virus titers comparable to those of PrV-Ka derived from the same cell line were found for RK13-UL31-P133A and RK13-UL31-D238A, showing functional complementation and pointing to unimpaired nuclear egress, as well as further virus maturation. In contrast, titers of PrV-ΔUL31 from RK13-UL31-D132A, RK13-UL31-D245A, RK13-UL31-C241-243A, RK13-UL31-K242A, and RK13-UL31-D249A were in the same range or only marginally higher than those derived from nontransgenic RK13 cells (Fig. 4), showing a striking defect in production of infectious virus progeny.
FIG 4.
One-step growth. Functional complementation was tested after infection of the cell clones expressing native pUL31 or mutated proteins with PrV-Ka or PrV-ΔUL31 at an MOI of 3. Infected cells and supernatants were harvested at 24 h p.i., and combined titers were determined on RK13-UL31 cells. Nontransgenic RK13 cells were used as a negative control. Shown are mean values of three independent experiments with the corresponding standard deviations. Statistically significant differences are indicated (ns, not significant [P > 0.05]; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Ultrastructural analyses.
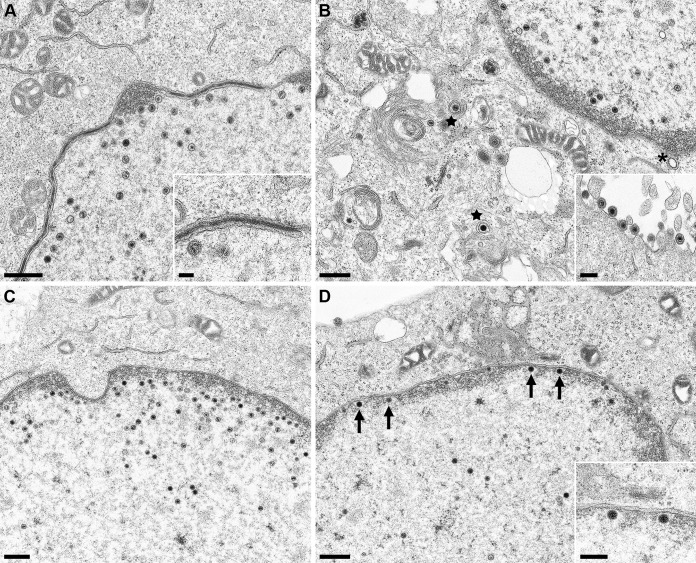
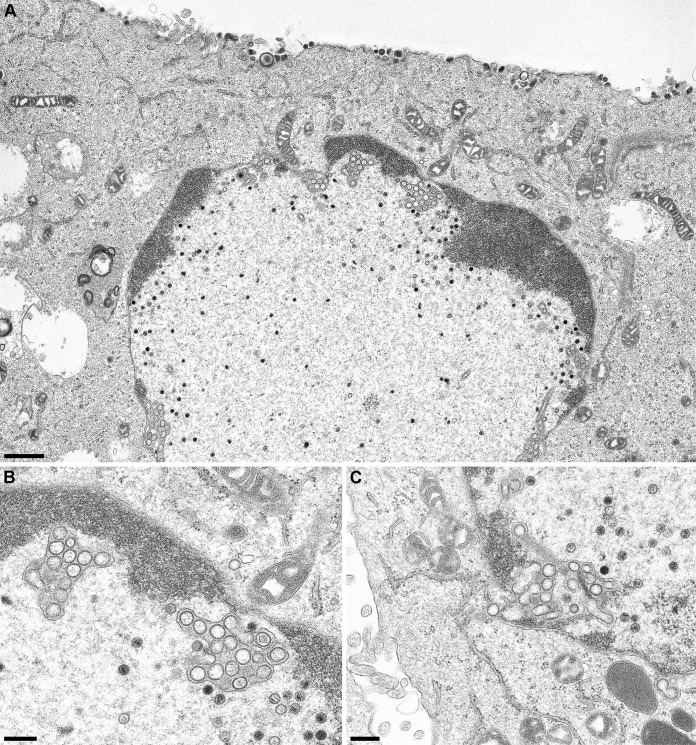
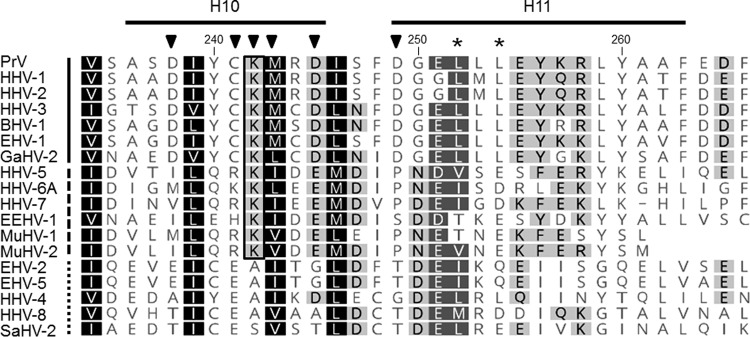
To test at which stage the formation of infectious progeny was impaired, pUL31 mutants that were unable to complement the defect of PrV-ΔUL31 were further investigated. To this end, RK13-UL31-D132A, RK13-UL31-C241-243A, RK13-UL31-K242A, and RK13-UL31-D249A cells were infected with PrV-ΔUL31 at an MOI of 1 and processed for electron microscopy 14 h p.i. as described previously (25). We also included RK13-UL31-D238A, which efficiently complemented the defect of PrV-ΔUL31 but is part of H10, which turned out to be important for capsid interaction. After infection of RK13-UL31-D132A (Fig. 5A), RK13-UL31-D245A (Fig. 5C), and RK13-UL31-D249A (Fig. 5D) capsids were only detectable in the nucleus. RK13-UL31-D132A (Fig. 5A) showed duplications of the nuclear membrane, where the membranes seemed to be tightly glued by electron-dense material resembling the NEC coat (Fig. 5A, inset). Membrane infoldings into the nucleus were detectable after infection of RK13-UL31-D245A (Fig. 5C) but no budding was observed despite the presence of DNA containing capsids. PrV-ΔUL31-infected RK13-UL31-D249A showed numerous nucleocapsids closely apposed at the INM (Fig. 5D, arrows, inset), but membrane curvature or budding did not ensue. In contrast, no impairment in nuclear egress was obvious in cells expressing pUL31-D238A (Fig. 5B). Mature and immature capsids in the nucleus, enveloped nucleocapsids, or capsids in the process of secondary envelopment (Fig. 5B, stars), as well as mature virions on the cell surface were detectable (Fig. 5B, inset). However, despite the obvious efficient transcomplementation of PrV-ΔUL31 by pUL31-D238A, empty vesicles resembling primary envelopes in the PNS were infrequently found (Fig. 5B, asterisk), indicating that efficient capsid uptake into primary envelopes might be affected. In contrast, in RK13-UL31-C241-243 (Fig. 6A and B) invaginations of the inner nuclear membrane filled with empty vesicles were obvious despite presence of numerous DNA containing capsids. The empty vesicles were similar in size and morphology to primary envelopes, indicating that the mutation abrogated capsid incorporation, whereas vesicle formation and scission apparently continued unimpaired. The same effect was evident when only K242 was changed to alanine (Fig. 6C). In both mutants, in addition to the virion-sized envelopes, tubular structures were also observed. Taken together, these data demonstrate that the charged lysine located in H10 is critical for incorporation of nucleocapsids into INM-derived primary envelopes. Alpha-helical domain H10, and specifically K242, is well conserved within the alpha- and betaherpesviruses (Fig. 7) but not in the gammaherpesviruses, pointing to a similar strategy for capsid-envelope interaction.
FIG 5.
Ultrastructural analyses. RK13 cells stably expressing pUL31-D132A (A), pUL31-D238A (B), pUL31-D245A (C), or pUL31-D249A (D) were infected with PrV-ΔUL31 (MOI = 1) and processed for electron microscopy at 14 h p.i. Representative images are shown. Mature capsids in the cytoplasm are marked by stars. A single vesicle in the PNS is indicated by an asterisk (B); nucleocapsids attached to the INM are highlighted by arrows (D). Scale bars: 500 nm in panels A to D, 250 nm in the insets in panels B and D, and 100 nm in the inset in panel A.
FIG 6.
Ultrastructural analyses of cells expressing pUL31 mutants defective for capsid incorporation. RK13-UL31-C241-243 cells (A and B) or cells expressing pUL31-K242 (C) were infected as described for Fig. 5. (B) Magnification image of the accumulations of vesicles in the PNS shown in the overview image (panel A). The same accumulations were evident after infection of RK13-UL31-K242 cells (C). Scale bars: 1 μm in panel A and 300 nm in panels B and C.
FIG 7.
Amino acid comparison of alpha-helical domains H10 and H11. Amino acid sequences of pUL31 homologs from PrV (AFI70796), HHV-1 (human herpesvirus 1; CAA32324), HHV-2 (CAB06756), HHV-3 (NP_040150), BHV-1 (bovine alphaherpesvirus 1; NP_045327), EHV-1 (equine alphaherpesvirus 1; AAT67286), GaHV-2 (gallid alphaherpesvirus 2; AAF66766), HHV-5 (CAA35412), HHV-6A (NP_042930), HHV-7 (AAC54699), EEHV-1 (elephant endotheliotropic betaherpesvirus; AGE10032), MuHV-1 (murine betaherpesvirus 1; ACE95567), MuHV-2 (AAF99152), EHV-2 (NP_042668), EHV-5 (AIU39595), HHV-4 (AAA45866), HHV-8 (AAB62601), and SaHV-2 (saimiriine gammaherpesvirus 2; Q01041) were aligned using ClustalW alignment (Geneoius version 10.0.9 [36]). Physicochemically similar residues conserved in all sequences aligned are shown by white letters on a black background, those conserved in >80% of the sequences are shown by white letters on a dark gray background, and those conserved in >60% of the sequences are shown in black letters on a gray background. The PrV pUL31 amino acid sequence is shown at the top, with the corresponding numbering and secondary structure elements plotted above the alignment. The lysine residue K242, which is conserved in the alpha- and betaherpesviruses, is boxed. Members of the alphaherpesviruses are marked by a solid line behind the virus abbreviation (left); betaherpesviruses are indicated by a dashed line and gammaherpesviruses by a dotted line. Residues located in H10 and H11 and changed in this study are indicated by a black triangle, and leucine residues mutated in the NES in a previous study (24) are marked by asterisks.
DISCUSSION
The recently solved structures of the NECs from different herpesviruses revealed a striking similarity despite only moderate amino acid sequence homology and shed further light on this viral vesicle formation and scission machinery (18, 19, 20, 21). Using a multimodal high-resolution imaging approach the coat formed by coexpression of PrV pUL31 and pUL34 in perinuclear vesicles could be visualized which revealed a protein layer with two hexagonal lattices, a membrane-proximal and a membrane-distal face, which could be assigned to the pUL34 and pUL31 moieties, respectively (22). These results showed that the major portion of the NEC components is involved in heterodimer, as well as higher oligomer formation, leaving only the membrane-distal end open for interaction with the capsid. To identify the capsid interaction interface, we mutated regions of the pUL31 NEC component predicted to be exposed at the membrane-distal end of the NEC. We targeted three different domains with potential interaction activity, including the conserved alpha-helical domain H10 (19), which has been suggested as the most likely capsid interaction domain (18).
Mutation of proline at position 133 (P133A), located in a small loop between alpha-helices H6 and H7 protruding at the membrane-distal end of the NEC (19), had no obvious effect on NEC formation or nuclear egress, and titers for PrV-ΔUL31 derived from the corresponding cell line were comparable to PrV-Ka. This was unexpected since proline residues are usually important structural elements but follows observations for several proline mutants in the pUL34 NEC component (26). In contrast, substitution of the neighboring aspartate by alanine (D132A) resulted in a nonfunctional protein despite efficient relocalization to the nuclear rim in the presence of pUL34, demonstrating that complex formation is not noticeably affected. However, in contrast to the punctated speckles found after coexpression of the wild-type proteins, phenotypically different flat tubular structures were evident, indicating that although pUL31-D132A is recruited to the INM by interaction with pUL34, alteration of the charged into a neutral residue might affect NEC oligomer topology. Ultrastructural analysis of PrV-ΔUL31 infected RK13-UL31-D132A showed nuclear membrane duplications which appear tightly attached by an electron-dense layer similar to the NEC coat (Fig. 5A, inset), which probably corresponds to the patch-like pUL31/pUL34 costaining pattern observed after coexpression (Fig. 2). Similar flat structures were evident after coexpression of pUL34 with pUL31-D245A, which is located at the end of H10, and the corresponding cell line did also not complement the defect of PrV-ΔUL31. Taken together, both residues are likely involved in membrane rearrangement by alteration of NEC-NEC contacts but seem not to play a role in capsid incorporation.
Cotransfection of pcDNA-UL31-D249A with pcDNA-UL34 resulted in a punctate pattern at the nuclear rim, but vesicle formation seemed to be reduced (Fig. 2). Ultrastructural analyses revealed an abundance of nucleocapsids in the nucleus, but neither budding stages at the nuclear envelope nor any viral particles in the cytoplasm or at the cell surface were found pointing to severely impaired nuclear egress. This is corroborated by the greatly reduced virus titers after infection with PrV-ΔUL31, which were only slightly higher than those found after infection of nontransgenic RK13 cells (Fig. 4). However, nucleocapsids were observed in close proximity to the INM, indicating that capsid transport to and docking at the INM takes place but that the induction of membrane bending is impaired. A similar phenotype was found in the absence of pUL25 (11) or after the expression of pUL31-NESPM, which carries two mutations (L252A/L254A) in the predicted nuclear export signal (24) (Fig. 7). D249 is part of this predicted NES, and all three residues are located in H11, which further supports the role of this alpha-helix in membrane remodeling.
Substitution of D238A, which is located at the beginning of the predicted capsid-binding alpha-helix H10 (18, 19), resulted in a wild-type-like colocalization pattern after coexpression with pUL34 and efficient complementation of PrV-ΔUL31. In electron microscopic images, besides normal budding and virus maturation stages, “empty” primary envelopes in the perinuclear space were infrequently detected, which are very rare in wild-type PrV infections. This indicates that capsid incorporation might be slightly compromised although this defect is barely reflected in viral titers. In contrast, the triple-mutant pUL31-C241-243A—in which C241, K242, and M243 were simultaneously altered to alanine—resulted in abrogation of complementation despite wild-type-like speckle formation after cotransfection with pUL34. Ultrastructural analyses revealed accumulations of primary envelopes devoid of capsids in the perinuclear space in large invaginations of the INM, despite obviously unimpaired capsid morphogenesis in the nucleus (Fig. 6A and B). This is exactly the phenotype to be expected when capsid incorporation is specifically blocked, while NEC formation and oligomerization resulting in membrane bending and scission proceed unabated. A comparable effect was observed when only K242 (Fig. 6C), located in the middle of this predicted capsid-binding alpha-helix, was altered identifying this amino acid as the key residue for capsid incorporation. In addition to the primary virion-sized empty vesicles tubular structures were also evident, indicating that the capsid supports membrane curvature into tightly fitting envelopes. The accumulation of primary envelopes in the PNS in the absence of capsid cargo in cells expressing these mutant pUL31 is reminiscent of cells overexpressing the NEC in the absence of virus infection (23). A similar phenotype has also been described in an analysis of a charge cluster mutant of HSV-1 pUL34 in which arginine residues at positions 158 and 161 were altered to alanine (27). This effect was attributed to deregulation of vesicle formation from the INM. However, whereas these INM-derived vesicles were pleomorphic and mostly larger than primary envelopes, the vesicles we observe are homogeneous and exhibit the size expected from capsid-less primary envelopes (see Fig. 6).
K242, as well as sequences surrounding this residue, are highly conserved in the alphaherpesviruses (Fig. 7) and encompass a predicted phosphorylation site (Y240 in PrV pUL31). It can be speculated that the phosphorylation status of this nearby tyrosine residue regulates NEC oligomerization after binding of the capsid. However, further mutational analyses are required to test this hypothesis.
Although a critical residue for nucleocapsid incorporation into nascent primary envelopes has now been identified, the interaction partner on the capsid side remains enigmatic. Binding of the NEC to the pUL25 component of the CVSC has been proposed (15, 16). This would explain the preferential incorporation of nucleocapsids into nascent primary envelopes over immature capsid forms, in particular during PrV infection (7). Electron tomographic analyses of purified primary enveloped particles support the notion that the CVSC is the major, but not the sole contact site for the primary envelope (8). However, other potential interactions, e.g., with pUL33 which is part of the terminase complex and was identified as interaction partner of pUL31 in varicella-zoster virus, might function as a hub linking cleavage/encapsidation with nuclear egress (28). Furthermore, the major capsid protein pUL19 may directly interact with the NEC (29). We plan to use our virus mutants with specific defects in capsid incorporation but unimpaired formation of primary envelopes in reversion analyses to uncover second site mutations which may give a lead to the capsid structure(s) interacting with the NEC.
It is also unclear where in the nucleus interaction between pUL31 and the nucleocapsid occurs. It has been suggested that pUL31 binds to intranuclear capsids within the nucleoplasm and that pUL31-decorated capsids are then recruited by membrane-bound pUL34. This mechanism would explain targeting of nucleocapsids to the INM but would require interaction of soluble pUL31 with the intranuclear capsid. H10 in PrV pUL31 which we identified as interacting with the capsid cargo is distal from pUL31 domains involved in interaction with pUL34. Thus, it is conceivable that it folds properly also in soluble pUL31 and might then be capable of interacting with the capsid without pUL34. Unfortunately, no crystal structure of pUL31 in isolation has been resolved due to instability of the non-complex-bound protein. Alternatively, the capsid may dock at pUL31 only after formation of the NEC. Both alternatives appear conceivable taking into account that the capsid-interacting region is spatially separated from the domains mediating pUL31-pUL34 contact. The formation of primary envelopes in the presence of numerous mature capsids but with a defect in in the interaction interface as shown here for pUL31-K242A points to the preformation of NEC before binding the capsid.
In summary, we show here that alpha-helix H10 of PrV pUL31 plays an important role in nucleocapsid incorporation into nascent primary envelopes. K242 was identified as a key residue in this process. Our data are congruent with the assumption that K242 is (part of) a direct capsid interaction surface located within helix 10 of the pUL31 NEC component which is critical for NEC-nucleocapsid binding.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney cells (RK13) and RK13 cells stably expressing pUL31 were cultivated in Dulbecco modified Eagle minimum essential medium supplemented with 10% fetal calf serum. Wild-type PrV strain Kaplan (PrV-Ka) (30) was propagated in RK13 cells, while PrV-ΔUL31 lacking UL31 was amplified in RK13-UL31 cells (31).
Generation of pUL31 mutants and stably expressing cell lines.
Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). pcDNA-UL31 (31) was used as the template with primers shown in Table 1. Correct mutagenesis was verified by sequencing. To generate stably expressing cell lines, RK13 cells were transfected with the pcDNA-UL31 constructs by calcium phosphate precipitation (32). At 2 days after transfection, the cells were transferred into media containing 0.5 mg/ml G418 (Invitrogen). After 10 to 14 days, G418-resistant cell clones were picked by aspiration. pUL31 expression was tested by indirect immunofluorescence using the polyclonal rabbit anti-pUL31 serum (31). Cell clones homogenously expressing pUL31 were further characterized.
TABLE 1.
Primers used for mutagenesis of PrV pUL31
| Primer | Sequence (5′ to 3′)a | Location in PrV-Ka (nt)b |
|---|---|---|
| UL31-D132A | GTG GCC GCG CGG GCA CCC TCG GAG CGC | 29197–29171 |
| UL31-P133A | GCC GCG CGG GAC GCA TCG GAG CGC GCC | 29200–29174 |
| UL31-D238A | GTC AGC GCG AGC GCC ATT TAT TGC AAG | 28882–28856 |
| UL31-C241-243A | GCG AGC GAC ATT TAT GCC GCG GCG CGG GAC ATC AGC | 28876–28841 |
| UL31-K242A | GCG AGC GAC ATT TAT TGC GCG ATG CGG GAC ATC | 28876–28844 |
| UL31-D245A | TGC AAG ATG CGG GCC ATC AGC TTC GAC | 28861–28835 |
| UL31-D249A | GAC ATC AGC TTC GCC GGG GAG CTG CTG | 28849–28823 |
Mismatches are underlined and in boldface.
Nucleotide (nt) positions correspond to GenBank accession number JQ809328; only “forward” primers are shown.
Laser scanning confocal microscopy.
For localization and colocalization studies pcDNA-UL31 and plasmids expressing the mutated pUL31 variants were either transfected singly or cotransfected with pcDNA-UL34 (25) into RK13 cells by calcium phosphate coprecipitation (32). Two days after transfection cells were fixed with 3% paraformaldehyde, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS), and incubated with the polyclonal rabbit anti-pUL31 (dilution 1:500) and a newly developed anti-pUL34 monoclonal antibody (4B10C, dilution 1:20). Bound antibody was detected by Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa 555-conjugated goat anti-mouse IgG (Invitrogen). Fluorescence images were acquired using a confocal laser-scanning microscope (SP5; Leica, Germany). Images were processed using ImageJ (33).
Western blot analysis.
Nontransgenic RK13 and transgenic RK13 cells expressing wild-type or mutated pUL31 were cultivated in 6-well cell culture dishes for 2 days and subsequently scraped into the medium. Cells were pelleted by low-speed centrifugation, washed twice with PBS, and resuspended in PBS-sodium dodecyl sulfate (SDS) sample buffer (0.13 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerin, 0,01% bromophenol blue, 10% mercaptoethanol). Genomic DNA was fragmented by sonication, and proteins were denatured by boiling for 3 min and separated on an SDS 10% polyacrylamide gel. After transfer onto a nitrocellulose membrane, the blot was incubated with the polyclonal rabbit anti-pUL31 serum (31). Monoclonal anti-alpha tubulin (Sigma) was used as a loading control. Bound antibody was detected by peroxidase-coupled goat anti-rabbit and anti-mouse antibodies, visualized by enhanced chemiluminescence (Bio-Rad Clarity Western ECL blotting substrate), and recorded in an image analyzer (Bio-Rad, Germany).
One-step growth kinetics.
To test for functional complementation, cell lines expressing either wild-type or mutated pUL31 were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 3. After 1 h at 4°C, the inoculum was replaced with fresh prewarmed medium, and cells were incubated at 37°C for 1 h. Thereafter, extracellular virus was inactivated by low-pH treatment (34), and cells were further incubated at 37°C. Cells and supernatant were harvested 24 h p.i., and progeny virus was titrated on RK13-UL31 cells. The mean values of three independent experiments and the corresponding standard deviations are shown. Statistical significance was evaluated using an unequal variance t (Welsh) test provided by GraphPad Prism 7 software.
Electron microscopy.
Cell lines stably expressing mutated pUL31 were infected with PrV-ΔUL31 at an MOI of 1 and processed for electron microscopy at 14 h p.i. as described previously (25).
ACKNOWLEDGMENT
This study was supported by the Deutsche Forschungsgemeinschaft (DFG ME 854/12-2).
We thank Karla Günther, Cindy Meinke, and Petra Meyer for technical help and Mandy Jörn for photographic assistance. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
REFERENCES
- 1.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J Virol 76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 5.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenz M, Vollmer B, Unsay JD, Klupp BG, Garcia-Saez AJ, Mettenleiter TC, Antonin W. 2015. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J Biol Chem 290:6962–6974. doi: 10.1074/jbc.M114.627521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klupp BG, Granzow H, Mettenleiter TC. 2011. Nuclear envelope breakdown can substitute for primary envelopment-mediated nuclear egress of herpesviruses. J Virol 85:8285–8292. doi: 10.1128/JVI.00741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newcomb WW, Fontana J, Winkler DC, Cheng N, Heymann JB, Steven AC. 2017. The primary enveloped virion of herpes simplex virus 1: its role in nuclear egress. mBio 8:e00825-17. doi: 10.1128/mBio.00825-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol 80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp BG, Granzow H, Karger A, Mettenleiter TC. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J Virol 79:13442–13453. doi: 10.1128/JVI.79.21.13442-13453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol 72:3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funk C, Ott M, Raschbichler V, Nagel CH, Binz A, Sodeik B, Bauerfeind R, Bailer SM. 2015. The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog 11:e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang K, Wills E, Lim HY, Zhou ZH, Baines JD. 2014. Association of herpes simplex virus pUL31 with capsid vertices and components of the capsid vertex-specific complex. J Virol 88:3815–3825. doi: 10.1128/JVI.03175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J Virol 85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeev-Ben-Mordehai T, Weberruss M, Lorenz M, Cheleski J, Hellberg T, Whittle C, El Omari K, Vasishtan D, Dent KC, Harlos K, Franzke K, Hagen C, Klupp BG, Antonin W, Mettenleiter TC, Grunewald K. 2015. Crystal structure of the herpesvirus nuclear egress complex provides insights into inner nuclear membrane remodeling. Cell Rep 13:2645–2652. doi: 10.1016/j.celrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lye MF, Sharma M, El Omari K, Filman DJ, Schuermann JP, Hogle JM, Coen DM. 2015. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J 34:2937–2952. doi: 10.15252/embj.201592651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walzer SA, Egerer-Sieber C, Sticht H, Sevvana M, Hohl K, Milbradt J, Muller YA, Marschall M. 2015. Crystal structure of the human cytomegalovirus pUL50-pUL53 core nuclear egress complex provides insight into a unique assembly scaffold for virus-host protein interactions. J Biol Chem 290:27452–27458. doi: 10.1074/jbc.C115.686527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bauerlein FJ, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grunewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A 104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passvogel L, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. 2015. Functional characterization of nuclear trafficking signals in pseudorabies virus pUL31. J Virol 89:2002–2012. doi: 10.1128/JVI.03143-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passvogel L, Janke U, Klupp BG, Granzow H, Mettenleiter TC. 2014. Identification of conserved amino acids in pUL34 which are critical for function of the pseudorabies virus nuclear egress complex. J Virol 88:6224–6231. doi: 10.1128/JVI.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roller RJ, Haugo AC, Kopping NJ. 2011. Intragenic and extragenic suppression of a mutation in herpes simplex virus 1 UL34 that affects both nuclear envelope targeting and membrane budding. J Virol 85:11615–11625. doi: 10.1128/JVI.05730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vizoso Pinto MG, Pothineni VR, Haase R, Woidy M, Lotz-Havla AS, Gersting SW, Muntau AC, Haas J, Sommer M, Arvin AM, Baiker A. 2011. Varicella-zoster virus ORF25 gene product: an essential hub protein linking encapsidation proteins and the nuclear egress complex. J Proteome Res 10:5374–5382. doi: 10.1021/pr200628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye GJ, Roizman B. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc Natl Acad Sci U S A 97:11002–11007. doi: 10.1073/pnas.97.20.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol 76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]