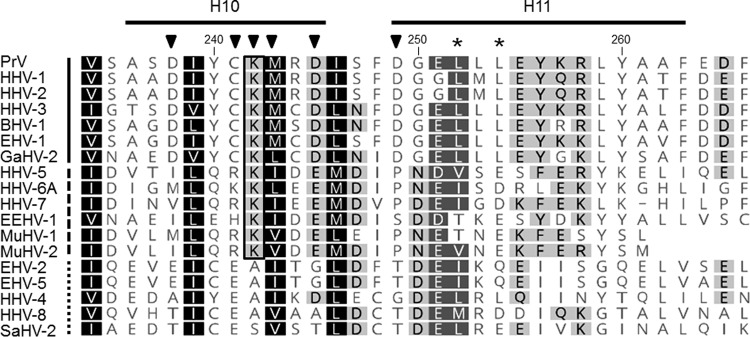
FIG 7.
Amino acid comparison of alpha-helical domains H10 and H11. Amino acid sequences of pUL31 homologs from PrV (AFI70796), HHV-1 (human herpesvirus 1; CAA32324), HHV-2 (CAB06756), HHV-3 (NP_040150), BHV-1 (bovine alphaherpesvirus 1; NP_045327), EHV-1 (equine alphaherpesvirus 1; AAT67286), GaHV-2 (gallid alphaherpesvirus 2; AAF66766), HHV-5 (CAA35412), HHV-6A (NP_042930), HHV-7 (AAC54699), EEHV-1 (elephant endotheliotropic betaherpesvirus; AGE10032), MuHV-1 (murine betaherpesvirus 1; ACE95567), MuHV-2 (AAF99152), EHV-2 (NP_042668), EHV-5 (AIU39595), HHV-4 (AAA45866), HHV-8 (AAB62601), and SaHV-2 (saimiriine gammaherpesvirus 2; Q01041) were aligned using ClustalW alignment (Geneoius version 10.0.9 [36]). Physicochemically similar residues conserved in all sequences aligned are shown by white letters on a black background, those conserved in >80% of the sequences are shown by white letters on a dark gray background, and those conserved in >60% of the sequences are shown in black letters on a gray background. The PrV pUL31 amino acid sequence is shown at the top, with the corresponding numbering and secondary structure elements plotted above the alignment. The lysine residue K242, which is conserved in the alpha- and betaherpesviruses, is boxed. Members of the alphaherpesviruses are marked by a solid line behind the virus abbreviation (left); betaherpesviruses are indicated by a dashed line and gammaherpesviruses by a dotted line. Residues located in H10 and H11 and changed in this study are indicated by a black triangle, and leucine residues mutated in the NES in a previous study (24) are marked by asterisks.