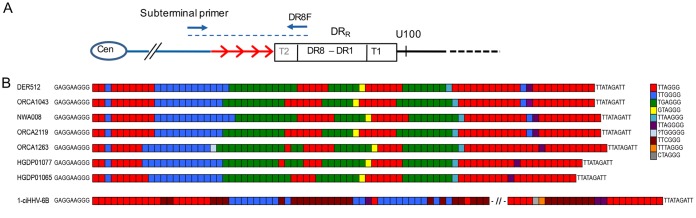
FIG 6.
Characterization of ciHHV-6B integration sites. (A) Diagram showing the locations of PCR amplicons used to characterize the chromosome–ciHHV-6B junctions. The red arrows represent TTAGGG and degenerate repeats; blue arrows, primers used to amplify the chromosome–HHV-6 junction; blue dashed line, chromosome junction amplicon used for sequence analysis. (B) Diagram showing the similarity of the TTAGGG and degenerate-repeat (see color key on the right) interspersion patterns in the chromosome–HHV-6 junctions from individuals with group 3 ciHHV-6B genomes (DER512 to HGDP01065 [Fig. 3B]). These interspersion patterns are distinct from that of the chromosome junction fragment isolate from 1-ciHHV-6B (singleton in Fig. 3B). The sequence on the left of the repeats is from the chromosome subtelomeric region, and the sequence on the right is from the ciHHV-6B genome.