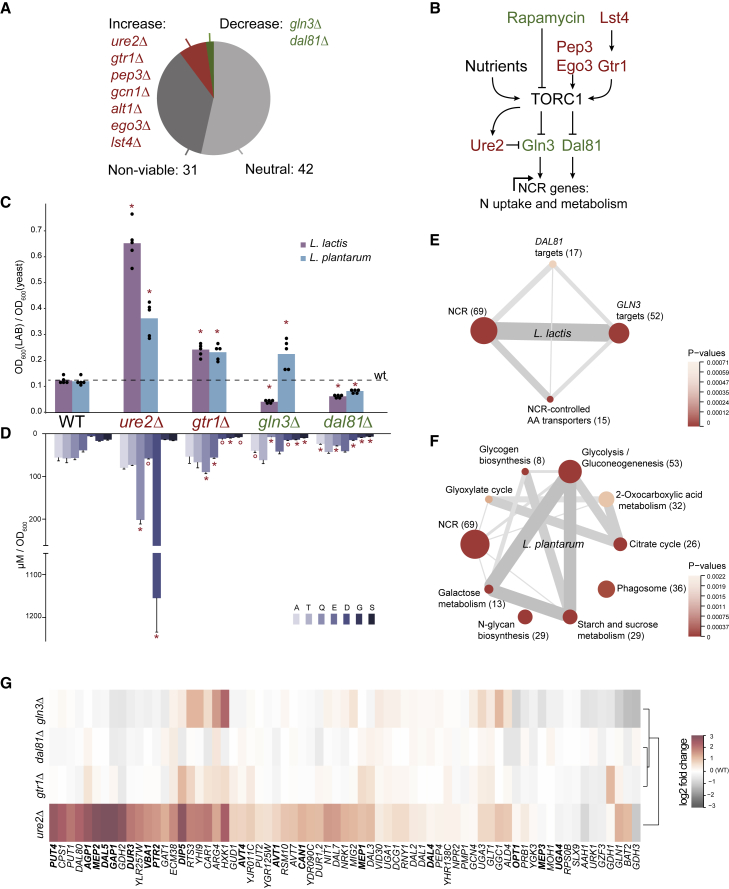
Figure 4.
Yeast Knockout Strains Indicate NCR-Sensitive Genes as Regulators of Interaction with LAB
(A) Effect of TORC1 pathway-related single-gene knockout strains on LAB growth (relative to the wild-type). See also Table S5.
(B) Interactions of NCR/TORC1 regulators that affect yeast-LAB interactions, as per literature review. Green color highlights proteins, of which the corresponding gene knockout downregulates the effect of yeast on LAB, and the red color shows effectors whose absence has a positive effect.
(C) Selected S. cerevisiae knockout mutants with altered effect on LAB growth (compared with the wild-type). ∗p < 0.01. n = 5 biological replicates.
(D) Concentrations of amino acids in exo-metabolome of knockout yeast strains. ∗p < 0.01, °p < 0.05. Data shown as mean ± SD (n = 4 biological replicates). See also Figure S7.
(E) Gene sets enriched for genes correlating with L. lactis growth (across the four selected knockout strains and the wild-type). Shown are groups with enrichment p < 0.01 (see the STAR Methods). Number of genes in each set is given in parentheses.
(F) Same as in (E) for L. plantarum. Shown are top ten non-redundant groups with enrichment p < 0.01.
(G) Expression of NCR genes (Ljungdahl and Daignan-Fornier, 2012) in selected knockout strains (log2 fold change relative to the wild-type, n=4 biological replicates). Amino acid and peptide transporters are shown in bold.