Figure 7.
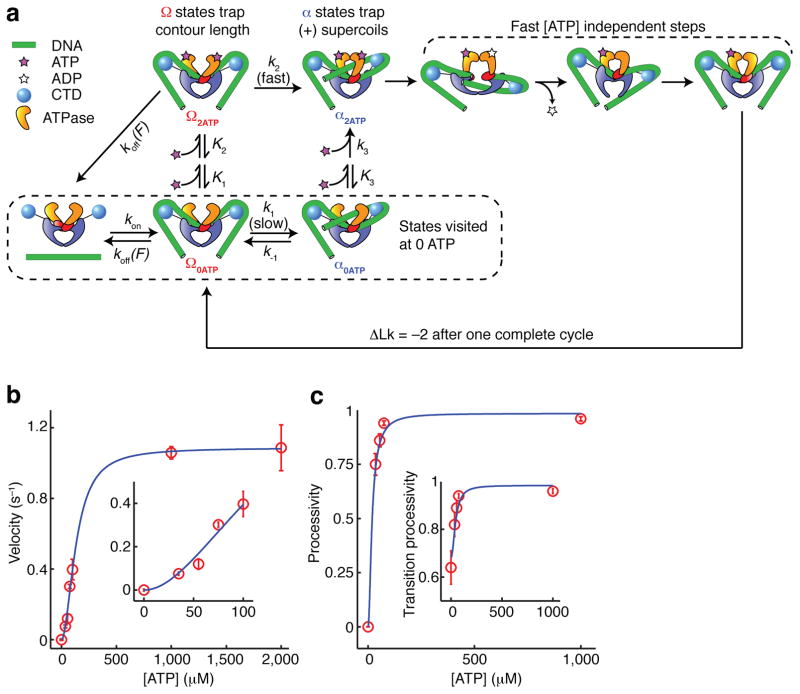
Branched kinetic model for structural transitions and ATP coupling in DNA gyrase. (See also Supplementary Figure 3.) (a) Diagram of allowed transitions in the model. Several ATP binding transitions are approximated as rapid equilibria with state-dependent affinities K1, K2, and K3. Rate constants are labeled for all other transitions. Some reverse rates are assumed to be negligible and are not shown. The dissociation rate koff(F) varies with the tension F in the DNA molecule; all other rates are tension-independent. Unlabeled arrows represent transitions of unknown kinetics but are fast relative to k2. In aggregate, they may occupy as much as 0.5 s of the cycle (Fig. 2b). (b) Overall supercoiling velocity (expressed in strand passages per second) as a function of [ATP], as measured experimentally and predicted from the kinetic model. Inset expands the low [ATP] region. (c) Processivity of the enzyme, defined as the probability of completing a forward step before dissociating, as a function of [ATP]. The transition processivity (inset) is defined as the probability of docking a T-segment before dissociating, and remains measurable at 0 ATP. Error bars indicate SEM.