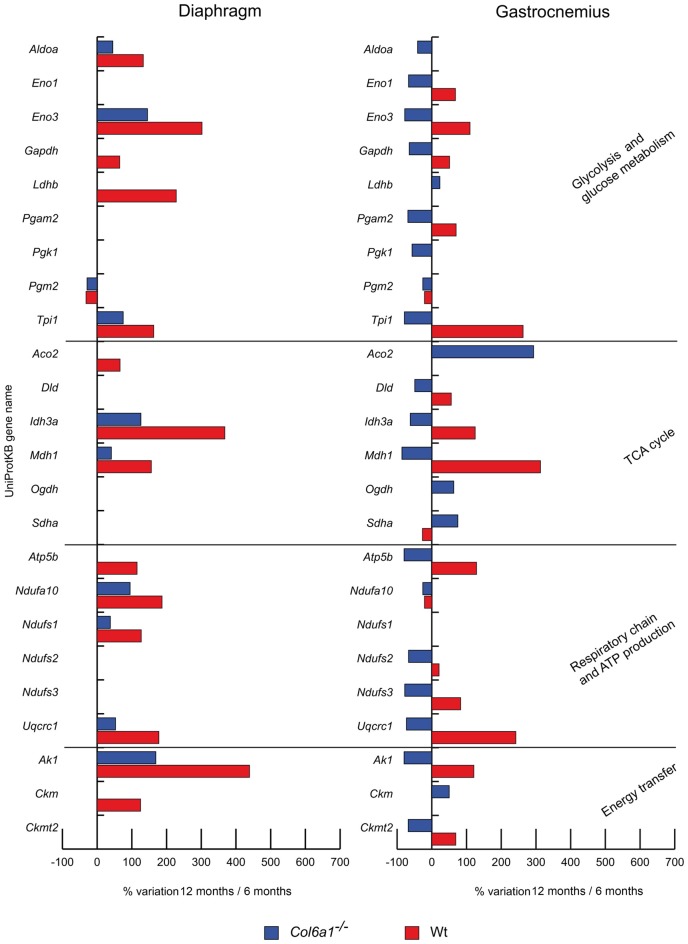
Figure 2.
Histograms of differentially expressed metabolic proteins in Col6a1−/− (blue bars) and Wt mice (red bars). Isoforms of proteins significantly altered (Student’s t-test, n = 4, p < 0.01) are expressed as % of spot volume variation in 12-month-old vs. 6-month-old mice (Aldoa: aldolase A; Eno1: enolase; Eno3: beta-enolase; Gapdh: glyceraldehyde-3-phosphate dehydrogenase; Ldhb: L-lactate dehydrogenase B; Pgam2: phosphoglycerate mutase; Pgk1: phosphoglycerate kinase 1; Pgm2: phosphglucomutase 2; Tpi1: triosephosphate isomerase 1; Aco2: aconitate hydratase; Dld: dihydrolipoamide dehydrogenase 1; Idh3a: isocitrate dehydrogenase alpha subunit; Mdh1: cytoplasmic malate dehydrogenase; Ogdh: oxoglutarate dehydrogenase; Sdha: succinate dehydrogenase; Atp5b: ATP synthase subunit beta; Etfa: electron transfer flavoprotein subunit alpha; Ndufa10: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10; Ndufs1: NADH-ubiquinone oxidoreductase 75 kDa subunit; Ndufs2: NADH-dehydrogenase iron-sulfur protein 2; Ndufs3: NADH-dehydrogenase iron-sulfur protein 3; Uqcrc1: ubiquinol-cytochrome c reductase core protein I; Ak1: adenylate kinase 1; Ckm: creatine kinase; Ckmt2: mitochondrial creatine kinase S-type).