Figure 1.
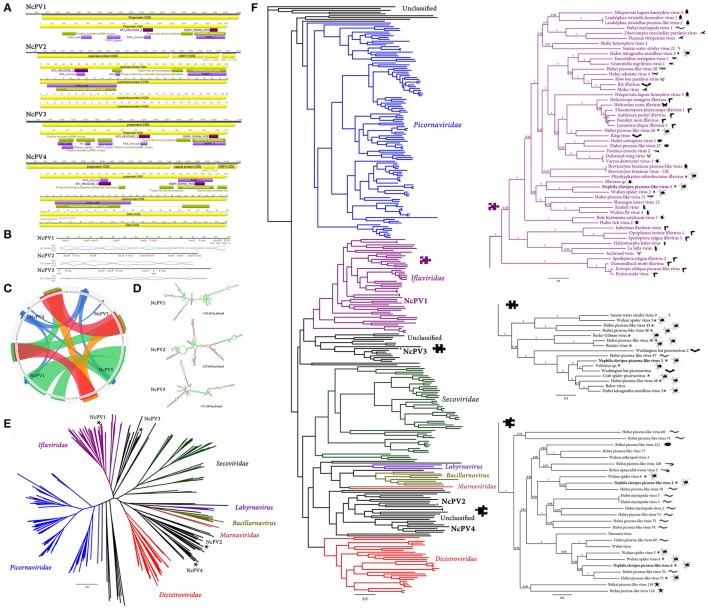
Nephila clavipes Picornavirales like viruses (A) Genome graphs depicting genomes and predicted gene products of N. clavipes picorna-like viruses. Pfam, PROSITE and Superfamily predicted domains (E-value ≤ 1e-5) are shown in purple, bordeaux and green, respectively. Predicted domain data is available in Supplementary Tables 1–4. (B) NcPV1-3 long A+U rich 5′ untranslated regions, presenting several UUUA motifs (loop) found typically in Picornavirales. Percentage of GC and AU content are expressed in blue and green line graphs, respectively. (C) Similarity levels of NcPV replicases expressed as Circoletto diagrams (Darzentas, 2010) based on BLASTP searches with an E-value of 1e-1 threshold. RPs are depicted clockwise, and sequence similarity is visualized from blue to red ribbons representing low-to-high sequence identity. (D) NcPV1-3 secondary structure of 5′ UTR, a RNA element predicted to function as internal ribosome entry site (IRES), allowing translation initiation in a cap-independent manner, as part of protein synthesis. (E) Maximum likelihood unrooted branched phylogenetic tree based in MAFFT alignments of predicted replicase proteins of Nephila clavipes picorna-like viruses (black stars) and related viruses. Families of viruses of the Picornavirales order are indicated by colors. Scale bar represents substitutions per site. (F) Rooted layout of the preceding phylogenetic tree. Magnifications of relevant regions of the tree are presented on the right and indicated by puzzle pieces. Reported hosts of viruses are represented by silhouettes. Branch labels represent FastTree support values. Complete tree showing tip species, host and virus assigned taxonomy labels are available as (Supplementary Figures 1–3). NcPV1-4, Nephila clavipes picorna-like virus 1–4.