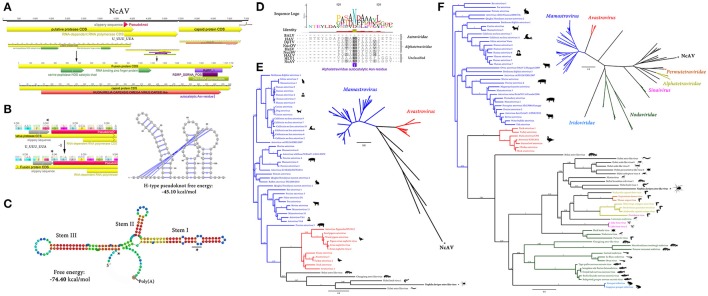
Figure 5.
Nephila clavipes Astroviridae like virus (A) Genome graphs depicting genome and predicted gene products of N. clavipes astro-like virus (NcAV). Pfam, PROSITE and Superfamily predicted domains (E-value ≤ 1e-5) are shown in purple, Bordeaux, and green, respectively. Functional domains determined by HHPRED are indicated in dark green. Predicted domain data is available in Supplementary Table 10. (B) NcAV predicted−1 ribosomal frameshifting is induced by a heptanucleotide “slippery sequence” of the form U_UUU_UUA, identical to the form of HIV1 (Retroviridae; Lentivirus), and Sindbis virus (Togaviridae; Alphavirus), followed downstream by a highly stable RNA H-type pseudoknot structural element. (C) NcAV presents a highly stable triple stem loop predicted RNA structure at its 3′ termini. Stem I of NcAV presents at an equilocal region a UCUU motif (black star) which in Human astrovirus 8 mediates PTB binding to the 3′ UTR, required for Astrovirus replication. (D) MAFFT alignment of the capsid like protein of NcAV and related Astroviridae, Alphatetreviridae, and unclassified astro-like viruses. Maturation of the capsid in alphatetraviruses is mediated by an autoproteolytic cleavage dependent of the presence of an Asn-570, which is at the cleavage site, and is present in NcAV, and Alphatetraviridae and unclassified astro-like viruses, but not in Astroviridae viruses. Beihai astro-like virus (BALV), Human astrovirus (HAV), Dendrolimus punctatus tetravirus (DpTV), Nudaurelia capensis omega virus (NucOV), Helicoverpa armigera stunt virus (HaSV), Hubei astro-like virus (HALV), Hubei leech virus 1 (HLV1). (E) Maximum likelihood unrooted branched phylogenetic tree based in MAFFT alignments of predicted RP protein, or Capsid protein (F) of Nephila clavipes astro-like virus (black stars) and related viruses. Genera of viruses of the Astroviridae family and related virus families are indicated by colors. Scale bar represents substitutions per site. Rooted layout of the preceding phylogenetic trees are also presented. Reported hosts of viruses are represented by silhouettes. Branch labels represent FastTree support values. Complete tree showing tip species, host, and virus assigned taxonomy labels are available as (Supplementary Figures 13–15).