Figure 1.
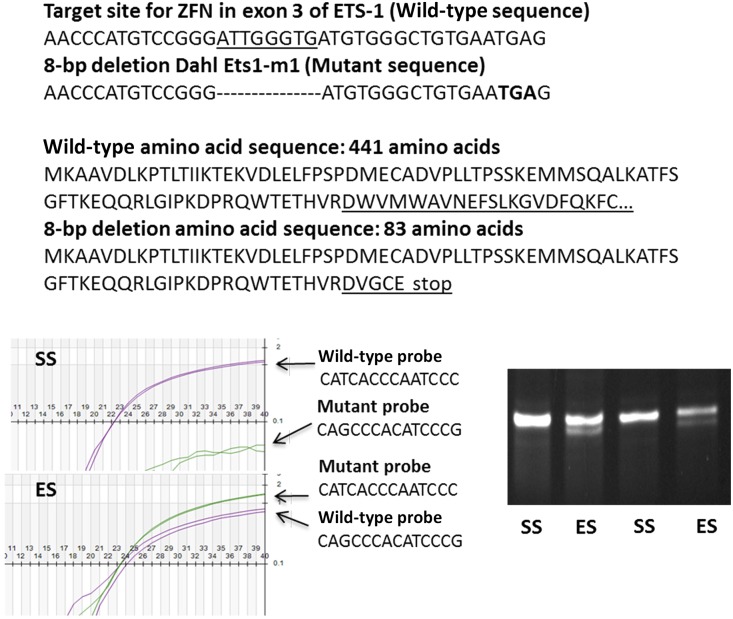
Genotyping confirmed the presence of a single mutated Ets-1 in ES rats. As shown, the 8-bp deletion was predicted to create a frameshift mutation, generating a premature stop codon (TGA) and a subsequent truncated, nonfunctional ETS-1 protein of only 83 amino acids. This deletion produced a nonfunctional mutant ETS-1 protein that lacked a critical transcription domain (amino acid residues 131–331) and DNA-binding domain (amino acid residues 331–440). Genotyping was confirmed using Taqman probes that recognized wild-type Ets-1 and the mutation (bottom left panel). The genotypes were determined as wild-type (homozygous SS) (upper left), heterozygous (ES) (lower left), or homozygous for the mutation, depending upon whether the samples were positive with only wild-type probes, positive with both wild-type and mutant probes, or positive with only mutant probes, respectively. During breeding, we did not find any rat pups homozygous for the mutation. To confirm the mutation in Ets-1 mRNA, total RNA was extracted from the rat kidney and the cDNA was amplified with PCR primers covering the target cDNA site; the resulting amplicons were separated on a 5%–10% polyacrylamide gel (bottom right). Wild-type (SS) had one band (100-bp, as predicted) and heterozygotes (ES) had two bands (100-bp and 92-bp, as predicted).