Abstract
Overexpression of the survivin gene contributes to tumorigenesis; it has been recognized as an important target for cancer therapy. In the present study, survivin expression was suppressed using recombinant plasmid mediated short hairpin RNAs (shRNAs) that were constructed to target exonic or intronic sequences of the survivin gene. In addition, a negative control shRNA was constructed. HeLa cells were transfected with specific shRNA constructs and the blocking efficiency of each shRNA was assessed at the mRNA and protein levels; and the five shRNA constructs with higher blocking efficiency were selected. Cell apoptosis was assessed by flow cytometry (FCM) following Annexin V-fluorescein isothiocyanate/propidium iodide double staining. Hoechst staining was used to detect the morphological diversity of the nuclei in apoptotic cells. The results demonstrated that survivin expression was effectively reduced by the transfection of shRNAs in HeLa cells. In addition, the apoptotic rates of the shRNA-treated groups were significantly increased compared with the negative control group according to the FCM results. The nuclei of HeLa cells exhibited apoptotic characteristics in the shRNA-treated groups as identified by Hoechst staining. Survivin-targeting shRNAs effectively downregulated the expression of the gene and markedly increased the apoptotic rate of HeLa cells. Data from the present study also indicated that the intron-specific shRNA demonstrate a high efficiency of inhibition of survivin expression and were able to induce cell apoptosis of HeLa cells through RNAi, potentially providing novel target sites for tumor therapy. In conclusion, the present study suggests that intron-specific blocking of survivin by RNAi may provide a tool for anticancer therapy.
Keywords: survivin, RNA interference, short hairpin RNA, apoptosis, intronic
Introduction
The survivin gene is located at the 17q15 region chromosomal region and contains four exons and three introns (1). Survivin was firstly separated from a human genomic library using the complementary DNA (cDNA) of effector cell protease receptor-1 (EPR-1) through hybridization screening (2). Survivin, containing only one baculovirus inhibitor of apoptosis protein repeat (BIR) domain, is the smallest member of the mammalian inhibitors of the apoptosis protein (IAP) family (2,3). Survivin is detectable during fetal development and is highly expressed in various malignant tumors, but is undetectable in adult terminal differentiation tissues (2,4–6). Survivin serves multiple functions in cell division, apoptosis, metastasis and angiogenesis (7). It has been reported that survivin acts as an intermediary between cell apoptosis and cell cycle checkpoints, serving an important role in reducing cellular apoptosis and regulating mitosis (8). Survivin suppresses apoptosis through inhibition of the terminal effectors caspase-3 and caspase-7, promoting the differentiation of tumor cells (6,9). Survivin expression peaks at the G2/M phase of the cell cycle, where it associates with mitotic spindle microtubules; its overexpression is required to protect cells from apoptosis during mitosis (10). It has previously been reported that the overall survival rate of patients that express high levels of survivin is decreased, and that survivin is an indicator of poor prognosis (11,12); the recurrence rate of these patients was increased and patients were not sensitive to radiotherapy and chemotherapy (13). Therefore, survivin may be a promising target for anticancer therapy (14).
RNA interference (RNAi) is the sequence-specific silencing of genes induced by endogenous or exogenous 21–23 nt double-stranded RNA in cells (15). RNAi is a feasible and effective method of inhibiting the endogenous expression of target genes, and has been widely employed in the functional studies of genes in vivo and in vitro (16,17). Two modified strategies of RNAi technology have been used to block gene expression in mammalian cells, including small interfering RNA (siRNA) and vector-mediated short hairpin RNA (shRNA) (18). Compared with shRNA vectors, siRNAs are more readily synthesized and delivered into cells; however, siRNA-mediated gene silencing is transient and siRNAs are susceptible to RNase degradation (19–21). shRNAs, on the other hand, can be used for long-term gene silencing and can also be propagated indefinitely (22,23). Once delivered into cells, shRNAs are cleaved into active siRNAs by Dicer, which then induce the homology-dependent degradation of cognate mRNA (24).
In the present study, the vector-derived shRNA technique was utilized and the recombinant plasmid pGP-U6-GFP-Survivin-shRNA was constructed, using a range of different shRNAs with a U6 small nuclear RNA promoter. The shRNAs used were targeted against different sites of the survivin gene, including exonic and intronic sequences, and were transfected into HeLa cells. The influence of survivin expression and cell apoptosis were both investigated. The present study may aid the development of a theoretical foundation for gene therapy in cancer.
Materials and methods
Cells line and culture
The human cervical carcinoma HeLa cell line was provided by the Institute of Biochemistry and Molecular Biology of Guangdong Medical University (Zhanjiang, China). The HeLa cells were cultured in Dulbecco's Modified Eagles Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Zhejiang Tianhang Biotechnology Co., Ltd., Zhejiang, China), 100 U/ml penicillin, and 0.1 g/ml streptomycin (Beyotime Institute of Biotechnology, Shanghai, China) at 37°C in a humidified atmosphere containing 5% CO2.
Recombinant plasmid-based survivin-targeting shRNA design
A total of 6 shRNAs targeting the survivin gene (and 1 negative control) were designed to be homologous to the survivin mRNA sequence (GeneBank no. NM001168). shRNA1 was targeted at intron 1; shRNA2 was targeted at intron 2; shRNA3 and shRNA4 were targeted at intron 3; shRNA5 was targeted at exon 1 and shRNA6 was targeted at exon 4 (Table I). According to BLAST analysis, the nucleotide sequences of survivin-targeted did not exhibit any non-specific interactions with other mRNA transcripts. All nucleotide sequences were synthesized and inserted into the recombinant plasmid vectors pGP-U6-GFP-Survivin by Shanghai GenePharma Co., Ltd. (Shanghai, China). The company presented a negative control shRNA (shNC), which did not possess any complementary region with survivin mRNA.
Table I.
Sequences of survivin-shRNAs and negative control.
| shRNA | Sequence length | Number of nucleotides |
|---|---|---|
| shRNA1 | GGTGATGCTTACAACCTAA | 19 |
| shRNA2 | GGGAGAGAGAAGGTGCTAA | 19 |
| shRNA3 | GCTCATGCTTTCCTTGCTA | 19 |
| shRNA4 | GCATTGGGCGCTGATTCTT | 19 |
| shRNA5 | CCGCATCTCTACATTCAAGAA | 21 |
| shRNA6 | GCACCACTTCCAGGGTTTATT | 21 |
| shNC | GTTCTCCGAACGTGTCACGT | 20 |
shRNA, short hairpin RNA; shNC, negative control shRNA.
Transfection of the shRNAs into HeLa cells
Cells were seeded into 6-well plates in DMEM without antibiotics prior to transfection. When cells were at 60–70% confluence in monolayer, transfection of shRNA was performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol, with a non-transfected group acting as the blank control. The shRNA:lipid reagent was used at a ratio of 1:2; each shRNA was transfected into three marked wells, cultured in humidified incubator and the medium was changed with fresh medium each day.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
After a 48-h transfection, the total RNA of all groups was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. cDNA was synthesized from total RNA (1 µg) using the FastQuant RT kit (With gDNase) (Tiangen Biotech Co., Ltd., Beijing, China). qPCR was performed using the SYBR®Premix Ex TaqII kit (Takara Bio, Inc., Otsu, Japan) on LightCycler®480 (Roche, Switzerland). The human GAPDH gene served as an internal control. The primers used were: Survivin forward, 5′-TGACGACCCCATAGAGGAACA-3′ and reverse, 5′-CGCACTTTCTCCGCAGTTTC-3′; and GAPDH forward, 5′-GGGTGTGAACCATGAGAAGT-3′ and reverse, 5′-CAGTGATGGCATGGACTGTG-3′. The final qPCR volume was 20 µl, with 2 µl cDNA template and 0.4 µM of each primer. The qPCR parameters were as follows: Initial denaturation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and annealing and extension at 60°C for 20 sec. Results were analyzed in relation to GAPDH levels using the 2−ΔΔCq method (25). Each experiment was performed in triplicate.
Western blot analysis
The total protein was obtained from each group using cold radio immune precipitation lysis buffer (cat. no. P0013B, Beyotime Institute of Biotechnology) containing phenylmethanesulfonyl fluoride (PMSF, catalog no. ST506-2, Beyotime Institute of Biotechnology) and protease inhibitors after a 72-h transfection. Protein concentration was quantified using the bicinchoninic acid (BCA) method using the Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. The proteins (60 µg/lane) were concentrated by 5% SDS-PAGE, separated by 12% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane (cat. no. ISEQ00010, EMD Millipore, Billerica, MA, USA). The membrane was blocked using 5% skimmed milk in Tris-HCl Buffered Saline Tween-20 (TBST, 25 mM Tris-HCl, 125 mM NaCl, 0.1% Tween-20) at room temperature for 2 h with gentle agitation, and incubated overnight at 4°C with monoclonal anti-survivin (1:1,000; cat. no. 2808T; Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-GAPDH (1:1,000; cat. no. 2118S, Cell Signaling Technology, Inc.) antibodies. Following a wash with TBST, the membrane was incubated with mice anti-rabbit IgG-horseradish-peroxidase monoclonal antibodies (1:2,000; cat. no. 5127S, Cell Signaling Technology, Inc.) for 1 h at room temperature. The antibodies were diluted with 5% skimmed milk in TBST buffer. The protein bands were then visualized using ECL Chemiluminescence Detection kit (Beyotime Institute of Biotechnology), using GAPDH protein as reference. The density of the brands on the membrane was scanned with Canon Solution Menu EX (Canon, Zhanjiang, China) and analyzed with ImageJ 1.46 software (National Institutes of Health, Bethesda, MD, USA).
FCM analysis
A total of 4×105 HeLa cells were seeded in 6-well plants. All groups of cells were treated with EDTA-free trypsin (Beyotime Institute of Biotechnology) for 3 min after a 48-h transient transfection. Cells were then centrifuged at 425 × g at 4°C for 5 min and the supernatant was discarded. Cells were then washed with cold PBS and centrifuged at 425 × g at 4°C for 5 min, removed and supernatant discarded carefully, twice. Cells were re-suspended in 100 µl of 1X Binding Buffer, and 5 µl Annexin V-fluorescein isothiocyanate (FITC) and 5 µl propidium iodide (PI) staining solution (cat. no. A221-01/02, all Vazyme Biotech Co., Ltd., Nanjing, China) were added, the solution was mixed gently and incubated at room temperature for 10 min in darkness. Next, 400 µl of 1X Binding Buffer was added. The ratios of apoptotic cells were assessed using a Coulter EPICS XL Flow Cytometer using Expo32-ADC v. 1.2B software (Beckman Coulter, Inc., Brea, CA, USA).
Hoechst 33258 stain analysis
In order to access nuclear condensation by Hoechst 33258 staining, 2×105 HeLa cells per well were washed twice with 2 ml PBS 48 h after transfection. A total of 1 ml Hoechst 33258 reagents (Beyotime Institute of Biotechnology) were added to each well and cells were incubated at 37°C for 30 min in the dark. Hoechst 33258 reagents were then removed and the cells were washed with PBS three times, 5 min each time. Morphological changes of apoptotic cells were observed under an inverted fluorescence microscope and images were captured.
Statistical analysis
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) software was used for statistical analysis. The results were presented as the mean ± standard deviation (SD) of three independent experiments. One-way analysis of variance was used to perform statistical comparisons of the data. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of survivin mRNA following transfection of HeLa cells with shRNAs
Survivin mRNA expression was evaluated in HeLa cells by RT-qPCR following transfection with recombinant plasmid vector-mediated survivin shRNAs (Fig. 1). For all groups treated with survivin-shRNA, survivin mRNA expression was significantly reduced compared with the blank control and shNC (P<0.05). No significant differences were observed in survivin mRNA expression between the shNC and the blank control groups (P=0.09). However, the effect of the suppression for the group transfected with shRNA1 was not well enough compared with other groups, and therefore it was not used in the following experiments.
Figure 1.
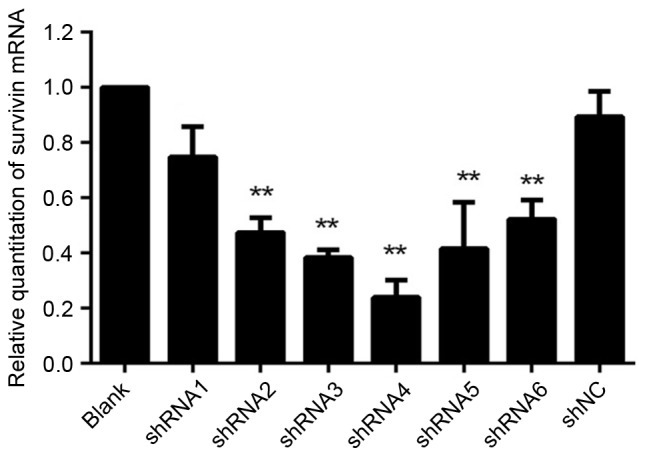
Quantitative representation of survivin mRNA levels. shRNAs were transfected into HeLa cells for 48 h and survivin mRNA levels were quantified by reverse transcription-quantitative polymerase chain reaction. Data shown are from three independent experiments. **P<0.01, compared with the blank control. shRNA, short hairpin RNA; shNC, negative control shRNA.
Expression of survivin protein following shRNAs transfection in HeLa cells
Survivin protein expression was examined by western blot analysis (Fig. 2). Survivin protein levels in all shRNA-treated groups were significantly reduced (P<0.05; Fig. 2B); however, no significant differences were observed between the shNC-treated and blank-control groups (P=0.40). The expression of survivin protein can not only be reduced by RNA1 extron-specific shRNAs but also inhibited by reduced intron-specific shRNAs in HeLa cells.
Figure 2.
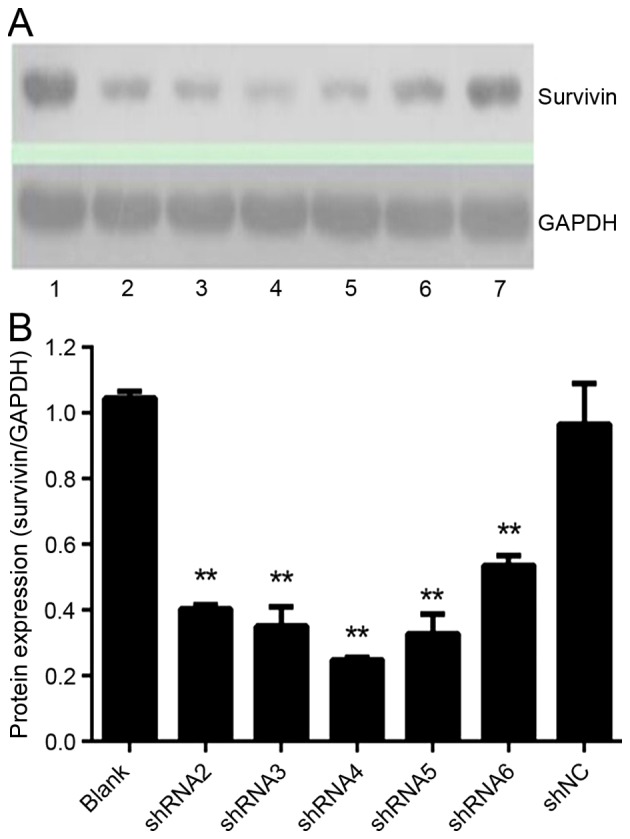
Effect of shRNAs on survivin protein expression. Protein levels were analyzed by western blot analysis after a 72-h transfection. (A) Bands of survivin and GAPDH were scanned. Lane 1, blank control; lane 2, shRNA2; lane 3, shRNA3; lane 4, shRNA4; lane 5, shRNA5; lane 6, shRNA6; lane 7, shNC. (B) The quantitative representation of survivin protein levels were determined by the density of the bands and one representation of three independent experiments was demonstrated, **P<0.01 vs. blank control. shRNA, short hairpin RNA; shNC, negative control shRNA.
Effect of shRNA treatment on apoptotic rates, assessed by flow cytometry (FCM)
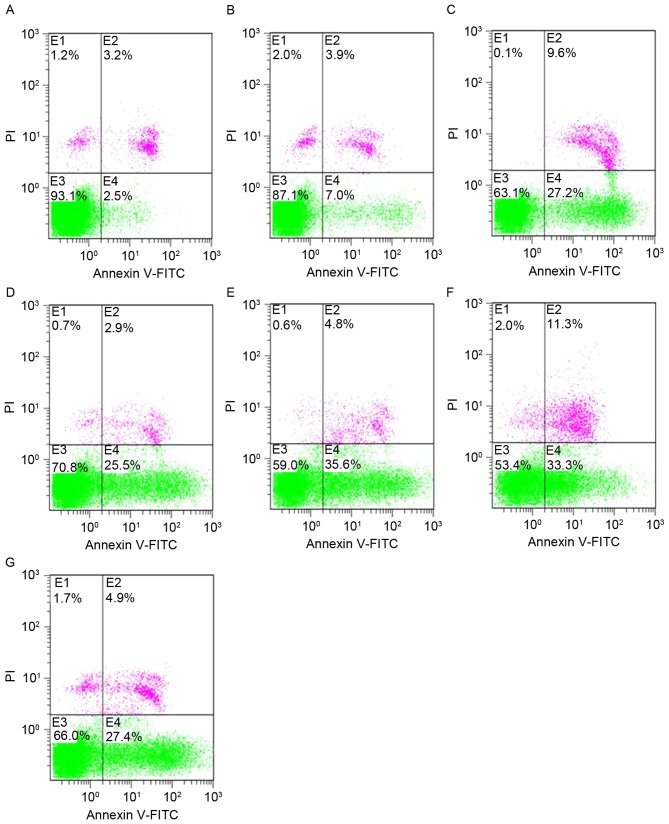
Cells transfected with shRNAs exhibited a significant increase in the level of apoptosis, as detected by FCM analysis via Annexin V-FITC/PI staining (Fig. 3; Table II). All data were presented as the mean ± SD of three independent experiments. The apoptosis rates were significantly increased in the shRNA-treated groups compared with shNC-treated and blank-control groups (P<0.05). No significant differences were observed between the shNC-treated and blank-control groups.
Figure 3.
Effect of shRNAs on cell apoptosis in HeLa cells by flow cytometry analysis. The E1 quadrant was the cell debris, the E2 quadrant was the necrotic cell group, the E3 quadrant was the surviving cell group and the E4 quadrant was the apoptotic cell group. (A) Blank control; (B) shNC; (C) shRNA2; (D) shRNA3; (E) shRNA4; (F) shRNA5; and (G) shRNA6. The increase in the number of apoptotic cells was marked. shRNA, short hairpin RNA; shNC, negative control shRNA; FITC, fluorescein isothiocyanate.
Table II.
Cell apoptosis rates after a 48-h transfection.
| Groups | Apoptosis rate, % |
|---|---|
| Blank control | 3.5±0.87 |
| shNC | 5.67±1.15 |
| shRNA-2 | 27.63±1.59a |
| shRNA-3 | 25.77±0.83a |
| shRNA-4 | 37.87±2.25a |
| shRNA-5 | 35.27±1.76a |
| shRNA-6 | 28.5±2.91a |
Data are expressed as the mean ± standard deviation of three independent experiments
P<0.01 vs. blank control. shRNA, short hairpin RNA; shNC, negative control shRNA.
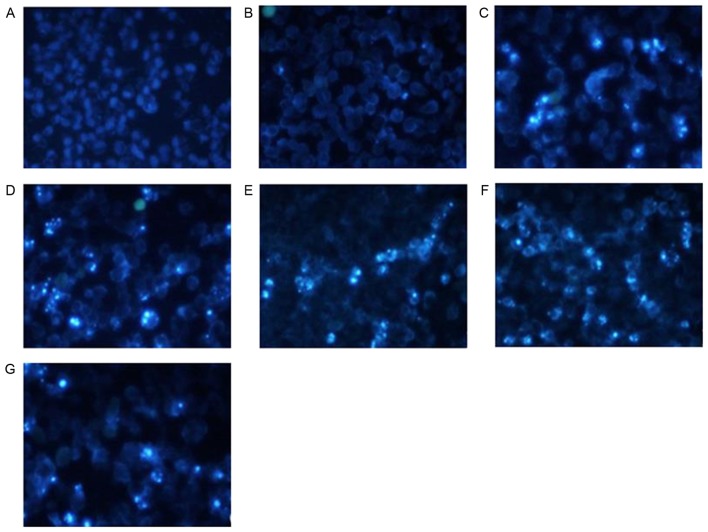
Effect of shRNA treatment on morphological changes of apoptotic cells by Hochest 33258 stain analysis
HeLa cells treated with shRNAs exhibited the following typical apoptotic changes following transfection (Fig. 4): Cell shrinkage, nuclear condensation and dense staining with some white color were observed under an inverted fluorescence microscope. Fluorescence staining in the group treated with shNC and the blank control group was light and uniform, and the number of apoptotic cells was markedly decreased.
Figure 4.
Effect of shRNA treatment on morphological changes of apoptotic cells by Hochest 33258 stain analysis. (A) blank control; (B) shNC; (C) shRNA2; (D) shRNA3; (E) shRNA4; (F) shRNA5; and (G) shRNA6. The cells treated with shRNAs exhibited changes typical of apoptotic cells following transfection: Cell shrinkage, nuclear condensation and dense stain with some white color. The fluorescence staining of the shNC and blank control groups was light and uniform.
Discussion
The capability of cells to escape apoptosis is one of the defined hallmarks of cancer (26,27). Multiple previous reports indicated that survivin serves a critical function during tumorigenesis by inhibiting apoptosis, accelerating the progress of mitosis and promoting the growth of tumor cells (28,29). High survivin expression is observed in the majority malignant tissues yet is absent in mature, healthy tissues (3), indicating it may represent a promising therapeutic target. Various strategies have been taken to inhibit the expression of survivin in cancer cells, including the use of antisense oligonucleotides (30), dominant-negative mutants (31), ribozymes (32,33) and anticancer vaccines (34). RNAi may be a powerful tool for cancer therapy (35). RNAi is efficient at lower concentrations for anticancer treatment and results in fewer side effects compared with other techniques (35,36). RNAi-mediated inhibition of survivin expression has been used to delay mitosis by causing chromosome misalignment and prometaphase accumulation in HeLa cells (19,37), and has successfully reduced survivin expression (26,38–40).
The vast majority of human genes are made up of introns. They have been considered to be ‘junk DNA’ because they are degraded following splicing (41–43). However, a previous study demonstrated that introns serve a critical role in transcriptional regulation (44). Introns are not only involved in the formation of microRNAs and small nucleolar RNAs, but also regulate alternative splicing and affect the mRNA level (45). In the present study, the shRNAs targeting exons and introns effectively inhibited the expression of survivin in HeLa cells and increased the apoptosis rate, inducing marked apoptotic morphological changes. This indicates that introns were not ‘junk DNA’.
The survivin gene contains four exons and three introns, and the survivin protein is the smallest member of the mammalian IAP family (2). To the best of our knowledge, few reports concerning the use of shRNA/siRNA targeted against introns to silence oncogenes exist, as the siRNA/shRNAs generated tend to target exons or promoters. As such, shRNAs in the present study were designed to block survivin expression by targeting exons and introns. A U6-promoter-mediated shRNA recombinant plasmid (pGP-U6-GFP-neo-Survivin) was utilized to inactivate survivin at multiple sites via multiple shRNA sequences. These plasmids were transfected into HeLa cells to evaluate the effectiveness of the survivin gene in vitro. All the shRNAs generated significantly downregulated expression of the target gene and protein, as assessed by RT-qPCR and western blot analysis. Notably, both FCM detection and Hochest 33258 staining revealed an increased degree of apoptosis in HeLa cells induced by survivin inactivation.
RNAi is an evolutionary conserved regulatory mechanism by which siRNAs induce the cleavage and degradation of homologous mRNA molecules specifically by transcriptional gene silencing (TGS) and post-TGS and (15). The shRNAs that target exons can decrease expression of survivin mRNA and protein in the cytoplasm; however, the intron-specific shRNAs cannot find the homologous mRNAs in cytoplasm, so they cannot have mediated their inhibitory role in this way. Previous reports have demonstrated that RNAi can suppress gene expression through TGS in plants (46,47), Drosophila melanogaster (48), Caenorhabditis elegans (49,50), and Schizosaccharomyces pombe (51). Many studies have revealed that RNAi-mediated TGS in mammalian cells does occur at the nuclear level, via RNA-directed DNA methylation, RNAi-mediated heterochromatin formation and the modification of histones (52–54). In the present study, we hypothesize that the shRNAs targeting introns may have blocked survivin gene expression through TGS.
Upon entry into the nucleus, shRNAs are processed by the microprocesor complex containing the RNase-III enzyme Drosha and DiGeorge Syndrome Critical Region 8 (55,56) into a form that can be recognized by exportin-5, a Ran-GTP-dependent nucleocytoplasmic transporter (57,58). The modified shRNAs are then exported from the nucleus into the cytoplasm by exportin-5. In the cytoplasm, the functional siRNAs are cleaved by Dicer in a complex with protein kinase R activator of transcription and Tar-RNA-binding protein. The siRNAs are loaded into Argonaute proteins (59,60), forming the pre-RNA-induced initiator of transcriptional gene silencing (pre-RISC) complex. With the assistance of partners, such as Dicer and Argonaute proteins (49,61), the pre-RISC complex translocates into the nucleus, where the RISC complex matures. Single stranded siRNAs directly bind to complementary DNA sequences, leading to DNA methylation, heterochromatin formation and the post-translational modification of histones, such as the trimethylation of Lysine 27 of histone H3 (H3K27me3), histone deacetylation, dimethylation of lysine 9 of histone H3 Lys9 (H3K9me2) (62,63). Several proteins may be involved in this model, including histone-lysine-methyltransferases, Histone-deacetylases, DNA methyltransferases, and histone protein 1.
Another alternative explanation for this experimental phenomenon is that exogenous siRNAs that target intron sequences may control alternative splicing, in a process similar to that of TGS (64). In this silencing pathway, the mature siRNAs directly recognize nascent pre-mRNAs by targeting intronic sequences, leading to heterochromatin formation (H3K9 dimethylation and H3K27 trimethylation) and DNA methylation by recruiting associated enzymes, subsequently modulating alternative splicing. As in TGS, Argonaute proteins, particularly Argonaute 1 (AGO1) and Dicer, are also involved in this mechanism (63). It is possible that once the siRNAs bind to the nascent pre-mRNAs with AGO1 and Dicer, they may cleave them directly to trigger transcriptional change. However, the other possible mechanisms cannot be excluded as explanations of the results of the present study. Whether the shRNAs target intron sequences to suppress survivin expression in vitro through the aforementioned mechanism remains unclear and will be revealed by further experiments.
In summary, the results of the present study demonstrate that the inhibition of survivin expression by shRNA may be a potential tool for cancer therapy and may provide further options for the design of interference target sites.
Acknowledgements
The authors would like to thank Professor Hou and Professor Huang for their guidance and Li Guodong for assistance with statistical analysis. The present study was supported by the Science and Technology Program of Higher Learning Institutions (Dongguan, China; grant nos. 200910815264 and 2012108102016).
References
- 1.Fulda S. Inhibitor of Apoptosis (IAP) proteins in hematological malignancies: Molecular mechanisms and therapeutic opportunities. Leukemia. 2014;28:1414–1422. doi: 10.1038/leu.2014.56. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 3.Wheatley S, McNeish IA. Survivin: A protein with dual roles in mitosis and apoptosis. Int Rev Cytol. 2005;247:35–88. doi: 10.1016/S0074-7696(05)47002-3. [DOI] [PubMed] [Google Scholar]
- 4.Soleimanpour E, Babaei E. Survivin as a potential target for cancer therapy. Asian Pac J Cancer Prev. 2015;16:6187–6191. doi: 10.7314/APJCP.2015.16.15.6187. [DOI] [PubMed] [Google Scholar]
- 5.Sanhueza C, Wehinger S, Bennett Castillo J, Valenzuela M, Owen GI, Quest AF. The twisted survivin connection to angiogenesis. Mol Cancer. 2015;14:198. doi: 10.1186/s12943-015-0467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15:2494–2516. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lladser A, Sanhueza C, Kiessling R, Quest AF. Is survivin the potential Achilles' heel of cancer? Adv Cancer Res. 2011;111:1–37. doi: 10.1016/B978-0-12-385524-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. Targeting survivin in cancer. Cancer Lett. 2013;332:225–228. doi: 10.1016/j.canlet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;113:4363–4371. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Duan N, Zhang C, Zhang W. Survivin and tumorigenesis: Molecular mechanisms and therapeutic strategies. J Cancer. 2016;7:314–323. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 13.Tran J, Master Z, Yu JL, Rak J, Dumont DJ, Kerbel RS. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc Natl Acad Sci USA. 2002;99:4349–4354. doi: 10.1073/pnas.072586399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coumar MS, Tsai FY, Kanwar JR, Sarvagalla S, Cheung CH. Treat cancers by targeting survivin: Just a dream or future reality? Cancer Treat Rev. 2013;39:802–811. doi: 10.1016/j.ctrv.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Chang S, Sun J, Zhu S, Pu C, Li Y, Zhu Y, Wang Z, Xu RX. Targeted microbubbles for ultrasound mediated short hairpin RNA plasmid transfection to inhibit survivin gene expression and induce apoptosis of ovarian cancer A2780/DDP cells. Mol Pharm. 2015;12:3137–3145. doi: 10.1021/mp500835z. [DOI] [PubMed] [Google Scholar]
- 17.Scholz C, Wagner E. Therapeutic plasmid DNA versus siRNA delivery: Common and different tasks for synthetic carriers. J Control Release. 2012;161:554–565. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling X, Li F. Silencing of antiapoptotic survivin gene by multiple approaches of RNA interference technology. Biotechniques. 2004;36(450–454):456–460. doi: 10.2144/04363RR01. [DOI] [PubMed] [Google Scholar]
- 20.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 21.Mohr SE, Perrimon N. RNAi screening: New approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3:145–158. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Q, van der Laan LJ, Janssen HL, Peppelenbosch MP. A dynamic perspective of RNAi library development. Trends Biotechnol. 2012;30:206–215. doi: 10.1016/j.tibtech.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Hu G, Luo J. A primer on using pooled shRNA libraries for functional genomic screens. Acta Biochim Biophys Sin (Shanghai) 2012;44:103–112. doi: 10.1093/abbs/gmr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Li QX, Zhao J, Liu JY, Jia LT, Huang HY, Xu YM, Zhang Y, Zhang R, Wang CJ, Yao LB, et al. Survivin stable knockdown by siRNA inhibits tumor cell growth and angiogenesis in breast and cervical cancers. Cancer Biol Ther. 2006;5:860–866. doi: 10.4161/cbt.5.7.2893. [DOI] [PubMed] [Google Scholar]
- 27.Igney FH, Krammer PH. Death and anti-death: Tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 28.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 29.Altieri DC. New wirings in the survivin networks. Oncogene. 2008;27:6276–6284. doi: 10.1038/onc.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 31.Yuan QZ, Wang CT, Mao YQ, Zhang P, Shi HS, Li ZY, Pan L, Yu DD, Leng F, Chen X, et al. Enhanced tumor radiosensitivity by a survivin dominant-negative mutant. Oncol Rep. 2010;23:97–103. [PubMed] [Google Scholar]
- 32.Pennati M, Binda M, Colella G, Folini M, Citti L, Villa R, Daidone MG, Zaffaroni N. Radiosensitization of human melanoma cells by ribozyme-mediated inhibition of survivin expression. J Invest Dermatol. 2003;120:648–654. doi: 10.1046/j.1523-1747.2003.12082.x. [DOI] [PubMed] [Google Scholar]
- 33.Pennati M, Binda M, De Cesare M, Pratesi G, Folini M, Citti L, Daidone MG, Zunino F, Zaffaroni N. Ribozyme-mediated down-regulation of survivin expression sensitizes human melanoma cells to topotecan in vitro and in vivo. Carcinogenesis. 2004;25:1129–1136. doi: 10.1093/carcin/bgh107. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Wang L, Wang H, Shang X, Niu W, Li J, Wu Y. A novel mimovirus vaccine containing survivin epitope with adjuvant IL-15 induces long-lasting cellular immunity and high antitumor efficiency. Mol Immunol. 2008;45:1674–1681. doi: 10.1016/j.molimm.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Chen Y, Ren J, Qu X. Small interfering RNA for effective cancer therapies. Mini Rev Med Chem. 2011;11:114–124. doi: 10.2174/138955711794519528. [DOI] [PubMed] [Google Scholar]
- 36.Joo MK, Yhee JY, Kim SH, Kim K. The potential and advances in RNAi therapy: Chemical and structural modifications of siRNA molecules and use of biocompatible nanocarriers. J Control Release. 2014;193:113–121. doi: 10.1016/j.jconrel.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 37.Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci. 2003;116:2987–2998. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- 38.Guo K, Song W, Gong Y, Hu S, Zhong W, Qiu W. Down-Regulation of survivin expression by siRNA suppresses proliferation and enhances chemosensitivity in human pancreatic cancer cell line Panc-1. In: Proceedings of the 2015 Seventh International Conference on Measuring Technology and Mechatronics Automation. IEEE, Nanchang. 2015:400–402. [Google Scholar]
- 39.Habib R, Akhtar J, Taqi M, Yu C, Zhang C. Lentiviral vector-mediated survivin shRNA delivery in gastric cancer cell lines significantly inhibits cell proliferation and tumor growth. Oncol Rep. 2015;34:859–867. doi: 10.3892/or.2015.4033. [DOI] [PubMed] [Google Scholar]
- 40.Zheng H, Tang C, Yin C. Oral delivery of shRNA based on amino acid modified chitosan for improved antitumor efficacy. Biomaterials. 2015;70:126–137. doi: 10.1016/j.biomaterials.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 43.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 44.Chorev M, Carmel L. The function of introns. Front Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lunghi M, Spano F, Magini A, Emiliani C, Carruthers VB, Di Cristina M. Alternative splicing mechanisms orchestrating post-transcriptional gene expression: Intron retention and the intron-rich genome of apicomplexan parasites. Curr Genet. 2016;62:31–38. doi: 10.1007/s00294-015-0506-x. [DOI] [PubMed] [Google Scholar]
- 46.Matzke MA, Primig M, Trnovsky J, Matzke AJ. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 48.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/S1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 49.Dernburg AF, Zalevsky J, Colaiácovo MP, Villeneuve AM. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- 50.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 53.Morris KV. RNA-mediated transcriptional gene silencing in human cells. Curr Top Microbiol Immunol. 2008;320:211–224. doi: 10.1007/978-3-540-75157-1_10. [DOI] [PubMed] [Google Scholar]
- 54.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 56.Mendez C, Ahlenstiel CL, Kelleher AD. Post-transcriptional gene silencing, transcriptional gene silencing and human immunodeficiency virus. World J Virol. 2015;4:219–244. doi: 10.5501/wjv.v4.i3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 58.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gagnon KT, Corey DR. Argonaute and the nuclear RNAs: New pathways for RNA-mediated control of gene expression. Nucleic Acid Ther. 2012;22:3–16. doi: 10.1089/nat.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53:245–257. doi: 10.1387/ijdb.082691av. [DOI] [PubMed] [Google Scholar]
- 62.Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burger K, Gullerova M. Swiss army knives: Non-canonical functions of nuclear Drosha and Dicer. Nat Rev Mol Cell Biol. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- 64.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]