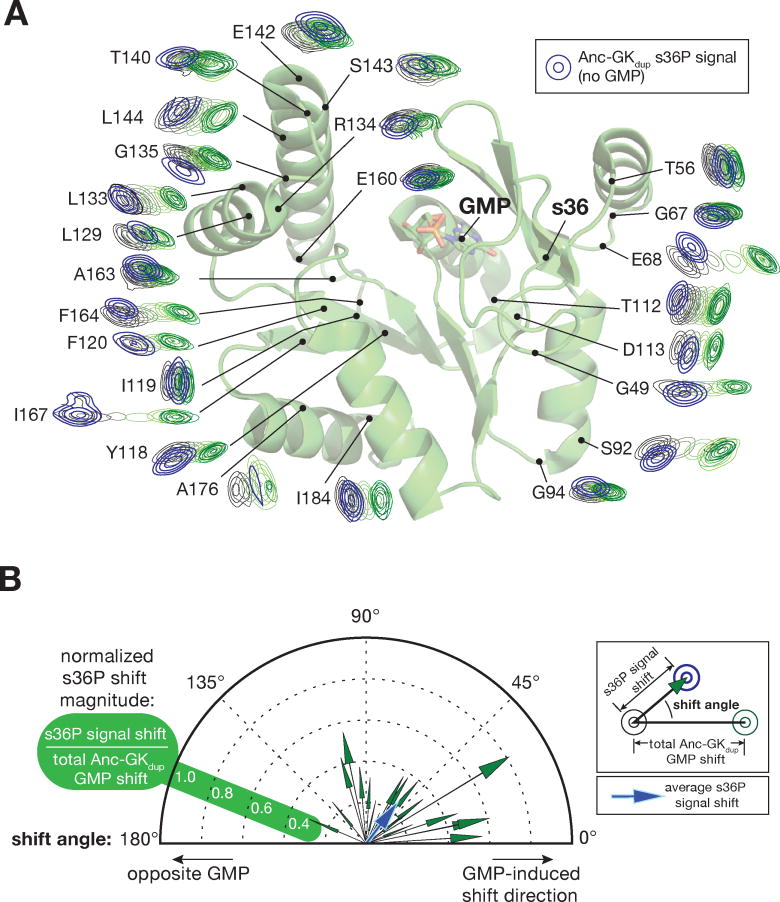
Figure 2.
Diverse conformational effects of GMP and the s36P PID mutation. A. The s36P mutation induces complex chemical shift perturbations. GMP binding and s36P induced chemical shift perturbations are shown mapped onto the GK structure. Assigned signals from Anc-gkdup and Anc-gkdup s36P residues distant from the GMP binding and mutation sites were taken from the HSQC spectra, rotated so that the Anc-gkdup signal without GMP is on the left and the GMP saturated signal is on the right, and placed on their position on the structure of GMP-bound yeast GK enzyme (PDB 1EX7). The HSQC signals from Anc-gkdup s36P are shown in blue. The full spectra with Anc-gkdup s36P overlaid are shown in Supporting Information. B. Comparison of chemical shift perturbations induced by the s36P mutation and GMP binding. Each vector represents the chemical shift perturbation of a single residue induced by the s36P mutation, relative to the shift induced by saturating concentrations of GMP. Signals from each residue shown in (A) were converted into vectors with angle defined by the angle between the s36P- and GMP-induced shifts (0° represents a shift in the same direction as GMP), and length defined by the ratio of the s36P- and GMP-shift magnitudes. Because the s36P induced shift magnitudes are normalized, residues with smaller absolute shifts may yield vectors of greater magnitude than those with larger absolute shifts. A vector representing the average shift is shown in blue.