Figure 4.
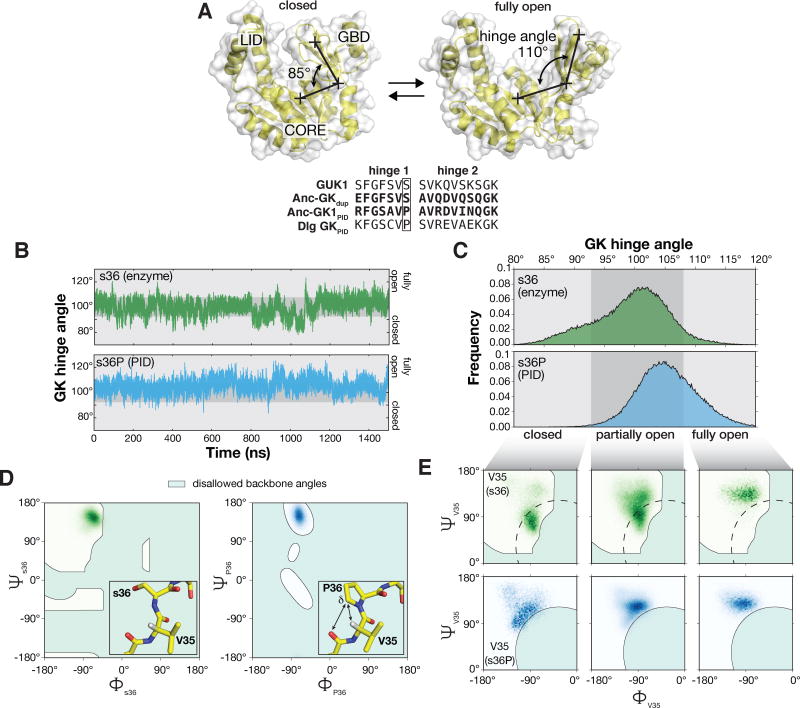
The s36P mutation preferentially destabilizes V35 peptide backbone conformations associated with the closed GK. A. Interconversion between closed and open GK. The three GK subdomains are shown: LID, CORE, and guanosine binding domain (GBD). The GK hinge angle is shown connecting crosses specifying the center-of-mass of the GBD, hinge residues, and CORE subdomains. Structures are from PDB 1EX7 (closed) and 1EX6 (chain A; open). An alignment of the hinge regions from the ancestral GKs are shown with those from the yeast GK enzyme (GUK1) and the extant GKPID from Discs large (Dlg GKPID). The s36P mutation is boxed. B. GK hinge angle as a function of molecular dynamics simulation time for GK s36 and s36P. Simulations were performed with structures from PDBs 1EX6 chain A (s36) and 4F4J (s36P). The s36 protein sampled hinge angles ranging from 78° – 123° during the simulation while s36P sampled angles from 83° – 126°. Closed (80° – 93°), partially open (93° – 108°), and open (108° – 120°) conformations are highlighted. C. Probability distribution functions for s36 and s36P GK hinge angles during the 1.5 μs molecular dynamics simulations shown in panel (B). D. Comparison of the backbone dihedral angles sampled by residue 36 (serine, left panel; proline, right panel) during 1.5 μs molecular dynamics simulations. The s36P mutation does not affect the conformations sampled by residue 36. Shaded areas represent disallowed angles for serine in the left panel and for proline in the right panel. Inset: protein structure near the s36P mutation. Interactions between the proline (s36P) δ carbon, valine 35’s (V35) α hydrogen, and serine 34’s (S34) carbonyl oxygen are shown. Structures are from PDB 1EX6 (chain A) and 4F4J (chain A).