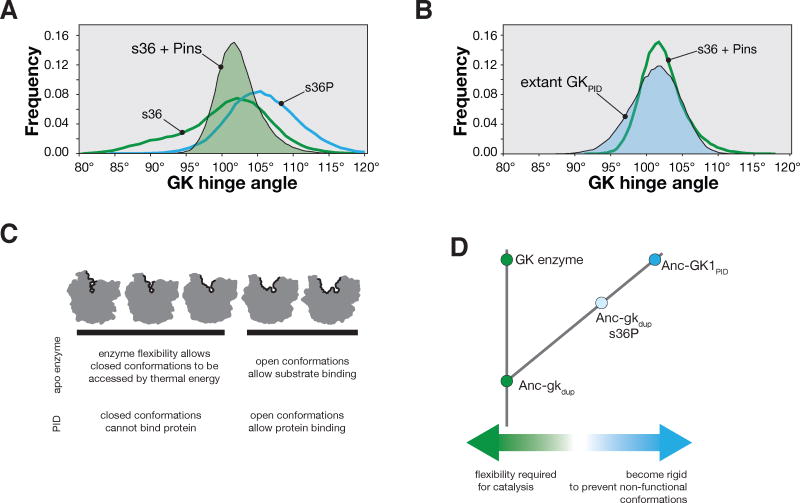
Figure 5.
The role of conformational flexibility in GK protein interaction domain evolution. A. Probability distribution function of s36 GK hinge angles when Pins is docked into its latent protein binding site during a 1.5 μs molecular dynamics simulation. The protein sampled hinge angles ranging from 90° – 119°. The distribution functions for s36 (without Pins) and s36P from Figure 4B are shown for comparison. B. Probability distribution function of the hinge angles from the extant GKPID from PSD-9527 during a 1.5 μs molecular dynamics simulation. The distribution function for the s36 GK enzyme with Pins docked (from panel A) is shown for comparison. C. Conformational flexibility model for the birth of the GK protein interaction domain family. The apo enzyme is able to access both open and closed conformations because of flexibility required for catalysis. The enzyme harbors a protein-binding site, but protein can only bind to a subset of conformations–thereby introducing an entropic penalty to binding. D. Diverging flexibility in GK enzyme and protein interaction domains. The ancestral enzyme (Anc-gkdup) is flexible, which allowed it to function catalytically, but prevented Pins binding. Extant enzymes retain these characteristics. A single mutation (s36P) significantly reduced GK flexiblity, abolishing catalytic activity but allowing the Pins binding site to become active by reducing the entropic barrier to binding. Binding was enhanced by mutations that further reduced the flexibility of AncGK1PID.