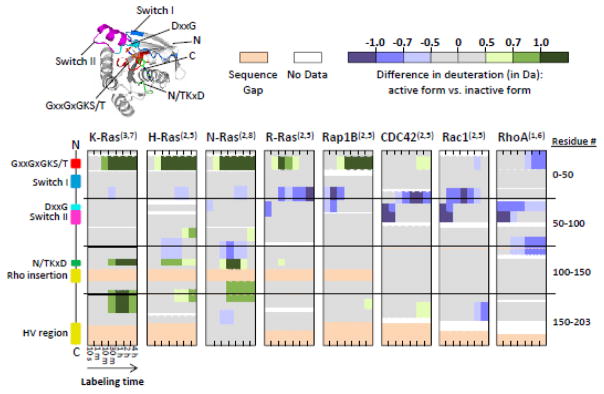
FIGURE 3.
Raw difference (in Da) between active, GMPPNP-bound and inactive, GDP-bound Ras proteins. From top to bottom, each visual shows the protein of interest from N- to C-terminus, with labeling time increasing from left to right. The superscript numbers in parentheses to the right of each protein name indicate the number of biological replicates (uniquely expressed and purified protein preparations) and the total number of labeling replicates analyzed, respectively, for that particular protein. The visual on the far left indicates the locations of key structural motifs, which are also highlighted on the crystal structure of active K-Ras (PDB: 3GFT) in the upper left corner. Gaps in coverage indicate regions of protein for which there was no peptide coverage and therefore no data were generated. Sequence gaps, represented in light beige, indicate regions where at least one protein had an insertion of one or more amino acids that other proteins did not share.