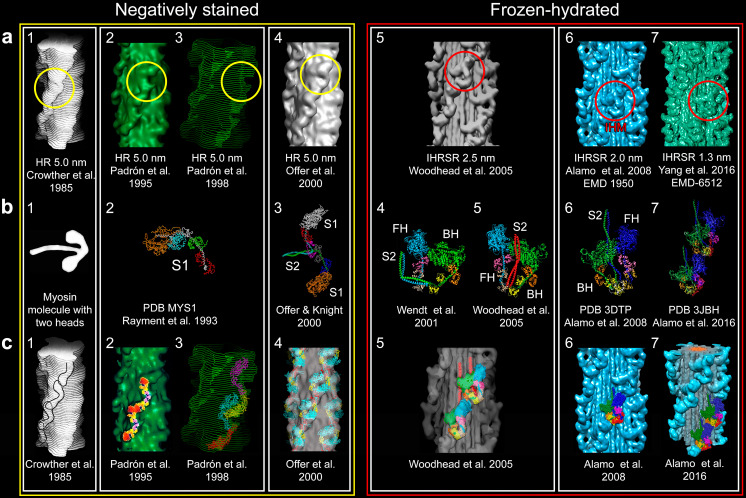
Fig. 4.
For the elucidation of the tarantula thick filament structure three requisites were needed to be fulfilled: a a better resolution of 3D reconstructions from negatively stained isolated filaments (a1–4 resolution 5.0 nm) or frozen-hydrated (a5 2.5 nm, a6 2.0 nm and a7 1.3 nm) with enhanced 3D reconstruction and visualization techniques (HR helical reconstruction, IHRSR iterative helical real space reconstruction), b the myosin head structure (b2) (PDB MYS1) and improved two-heads (b3–7) models and (c) fitting approaches ranging from eye-fitting (c1), 2D fitting (c2), envelope 3D fitting (c3), density rigid 3D fitting (c4, 5) and flexible 3D fitting (c6, 7). The unambiguous interpretation that lead to the myosin interacting-heads motif (IHM) (Woodhead et al. 2005) (a5, red circle) in the relaxed tarantula thick filament, belonging to the Class I proposed by Squire et al. (2005) and Squire (2009), required a resolution higher that 2.5 nm achieved by the IHRSR technique, frozen-hydrated specimens (a5) and an asymmetric head-pair model (b4) (Wendt et al. 2001), but with the S2 properly positioned (b5). In retrospect, the motif (red circles) was present and could be picked out on the 5-nm resolution 3D maps using the right density cutoffs (yellow circles). Bare zone is at the top here as well as in Figs. 6, 8, 9, and 10. Myosin sub-fragment 1 (S1), sub-fragment 2 (S2), free head (FH), blocked head (BH). The following images were reproduced with permission as follows: a1, c1 Crowther et al. (1985) and the Journal of Molecular Biology, a2, c2 Padrón et al. (1995) and the Journal of Structural Biology, c4 Offer et al. (2000) and the Journal of Molecular Biology, a5, b5, c5 Woodhead et al. (2005) and Nature, and c7 Alamo et al. (2016) and the Journal of Molecular Biology