Abstract
Background
DNA methylation is affected by the activities of the key enzymes and intermediate metabolites of the one-carbon pathway, one of which involves homocysteine. We investigated the effect of the well-known genetic variant associated with mildly elevated homocysteine: MTHFR 677C>T independently and in combination with other homocysteine-associated variants, on genome-wide leukocyte DNA-methylation.
Methods
Methylation levels were assessed using Illumina 450k arrays on 9,894 individuals of European ancestry from 12 cohort studies. Linear-mixed-models were used to study the association of additive MTHFR 677C>T and genetic-risk score (GRS) based on 18 homocysteine-associated SNPs, with genome-wide methylation.
Results
Meta-analysis revealed that the MTHFR 677C>T variant was associated with 35 CpG sites in cis, and the GRS showed association with 113 CpG sites near the homocysteine-associated variants. Genome-wide analysis revealed that the MTHFR 677C>T variant was associated with 1 trans-CpG (nearest gene ZNF184), while the GRS model showed association with 5 significant trans-CpGs annotated to nearest genes PTF1A, MRPL55, CTDSP2, CRYM and FKBP5.
Conclusions
Our results do not show widespread changes in DNA-methylation across the genome, and therefore do not support the hypothesis that mildly elevated homocysteine is associated with widespread methylation changes in leukocytes.
Introduction
DNA methylation, an important epigenetic mechanism has gained interest in the field of cancer and aging over the last decade [1, 2]. DNA methylation is affected by the activities of the key enzymes and intermediate metabolites of the one-carbon pathway, one of which involves homocysteine (Hcy).
Our aim was to investigate the role of genetically defined Hcy levels on genome-wide DNA methylation. A number of earlier studies have reported a link between Hcy and DNA methylation [3]. In these studies, DNA methylation was quantified as a global measure, that represents the total methyl cytosine content of the DNA. In animal models, both diet- and genetically- induced elevated Hcy have been related to altered global methylation patterns in tissues of aorta, brain, liver and colon. In human subjects, global DNA methylation in blood was not consistently altered with elevated Hcy. The relationship between Hcy and methylation can be subject to substantial bias, given the strong relationship between several lifestyle factors, diseases and Hcy. A way to circumvent this bias is to use genetic factors determining Hcy concentrations as an instrument to study the relationship between Hcy and methylation. The use of genetically defined elevated Hcy eliminate the effects that are possibly caused by measurement errors, confounding and reverse causality. One of the most consistent genetic variants causing elevated Hcy is the MTHFR 677C>T (rs1801133), which explains 5.3% variance in Hcy [4]. Furthermore, we recently published 18 variants including MTHFR 677C>T to be robustly associated with Hcy [5]. The Genetic Risk Score (GRS) of these 18 Hcy-associated variants explained 5.9% variance in Hcy [5]. In the current study, we used MTHFR 677C>T independently and the combined weighted GRS of these 18 variants, to test whether genetically defined elevated Hcy concentrations are associated with DNA methylation changes in blood cells.
A number of studies [3] have examined the relationship between the MTHFR 677C>T variant and global DNA methylation in humans. In 5 studies, individuals with the MTHFR 677TT genotype were compared to those with the MTHFR 677CC genotype [6–10]. Lower global methylation in blood cells was observed in two studies [6, 7]. The remaining three studies showed no association in the lymphocyte or colonic tissue. All published studies until now had small sample sizes of less than 200. To the best of our knowledge, associations of genetically defined Hcy with site-specific CpG methylation on a genome-wide scale have not been done up to now. In order to investigate this, we analyzed DNA methylation data measured with the Infinium Illumina 450k arrays, in a large meta-analysis of 9,894 individuals comprising 12 cohorts. We hypothesize that genetically defined elevated Hcy is associated with altered DNA methylation.
Materials and methods
Study population
All participants provided a written informed consent, and each study was approved at the relevant organizations by their respective ethics review committees [RS, Institutional review board (Medical Ethics Committee) of the Erasmus Medical Center; LLS, Ethical committee of the Leiden University Medical Center; LL, Ethics committee of the University Medical Centre Groningen; NTR, Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Centre; CODAM, Medical Ethical Committee of the Maastricht University; MARTHA, “Departement santé de la direction générale de la recherche et de l'innovation du ministère” (Projects DC: 2008–880 & 09.576); EGCUT; Research Ethics Committee of the University of Tartu; F5L, Research ethics boards of the University of Toronto and the Ottawa Hospital Research Institute; FHS, IRB (institutional review board); KORA, Local Ethics Committee; LBC1921, Lothian Research Ethics Committee (Wave 1: LREC/1998/4/183); LBC1936, Multi-Centre Research Ethics Committee for Scotland (Wave 1: MREC/01/0/56), and the Lothian Research Ethics Committee (Wave 1: LREC/2003/2/29)].
The analyses comprised of large population with 9,894 participants from 12 cohorts of European ancestry. Most of the cohorts were part of either the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium [11] and/or Biobank-based Integrative Omics Studies (BIOS) consortium [12]. All participants provided a written informed consent for the DNA collection and its use for genetic analyses. Each study was approved at the relevant organizations by their respective ethics review committees. Cohort-specific characteristics are provided in the S1 Text and S1 Table.
MTHFR 677C>T and homocysteine-associated SNPs
18 independent Hcy-associated SNPs from our GWAS meta-analysis [5], were selected to assess the relationship between mildly elevated Hcy concentrations and genome-wide DNA methylation. The genotypes of these SNPs were extracted from the genotyping data. Cohort-specific details of the quality control and the SNP imputation methods are provided in the S2 Table.
DNA methylation assessment
Whole blood samples were collected from the participants for DNA extraction. The genomic DNA was bisulfite converted using the Zymo EZ-96 DNA-methylation kit (Zymo Research, Irvine, CA, USA). Methylation profiling was performed using the Infinium Illumina HumanMethylation 450k BeadChip arrays (Illumina Inc., San Diego, USA) according to the manufacturers’ protocol. Beta values from 0 to 1, which represent the percentage of methylation, were calculated from the extracted raw methylated (M) and unmethylated (U) probe intensities and a default alpha (α) of 100. This is defined by the formula of β = M/(M+U+α). Normalization was performed on these raw beta values using DASEN [13] or SWAN [14] methods. Poor quality probes were excluded based on the detection p-values mostly >0.01 in >5% of samples. Cohort-specific data preprocessing methods are provided in the Table 1. Global methylation levels per sample was calculated by the mean of all CpGs as well as mean according to CpG islands, shores, shelves or non-coding regions [15].
Table 1. Details of methylation 450k pre-processing.
Quality control, normalization and association model.
| PROBES EXCLUSION | SAMPLE EXCLUSION | NORMALIZATION | ASSOCIATION MODEL | META-ANALYSIS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sr. No. | Cohorts | Detection p-value criteria | Cross-reactive & polymorphic | XY | Final | Criteria (Method) | Background correction | Dye bias correction | Normalization method | WBC counts | Technical covariates | Additional adjustments | Probe exclusion |
| CHARGE consortium | Probes with SNPs at SBE & probes with improper binding [16], probes that were absent in 8 or more studies, cis-probes with <5 Mb distance from Hcy-SNPs |
||||||||||||
| 1 | RS-III | >0.01 in >5% samples | Included | Excluded | 463,456 | Sample Call Rate >99%, poor bisulfite conversion, failed chromosome X & Y clustering | Yes | No | DASEN | Measured | Array, array position | No | |
| 2 | LBC1921 | >0.01 in >5% samples | Included | Included | 446,851 | >0.01 det. p-value in >5% probes, poor bisulfite conversion | Yes | No | None | Measured | Array, array position, plate, hybridization date | No | |
| 3 | LBC1936 | >0.01 in >5% samples | Included | Included | 446,851 | >0.01 det. p-value in >5% probes, poor bisulfite conversion | Yes | No | None | Measured | Array, array position, plate, hybridization date | No | |
| 4 | KORA | >0.01 | Included | Included | 441,487 | >0.01 det. p-value in >20% probes | Yes | Yes | BMIQ | Imputed | Array, array position | No | |
| 5 | FHS | None | Included | Included | 485,512 | Mismatched sex, outliers based on principal components (PCs) | Yes | No | DASEN | Imputed | Array, array position, PCs | No | |
| BIOS consortium | |||||||||||||
| 6 | RS | >0.01 in >5% samples | Included | Excluded | 419,937 | Poor bisulfite conversion | Yes | No | DASEN | Imputed | Array, array position | No | |
| 7 | LLS | >0.01 in >5% samples | Included | Excluded | 419,550 | Poor bisulfite conversion | Yes | No | DASEN | Imputed | Array, array position | No | |
| 8 | LLD | >0.01 in >5% samples | Included | Excluded | 420,591 | Poor bisulfite conversion | Yes | No | DASEN | Imputed | Array, array position | No | |
| 9 | NTR | >0.01 in >5% samples | Included | Excluded | 420,341 | Poor bisulfite conversion | Yes | No | DASEN | Imputed | Array, array position | No | |
| 10 | CODAM | >0.01 in >5% samples | Included | Excluded | 410,042 | Poor bisulfite conversion | Yes | No | DASEN | Imputed | Array, array position | No | |
| Other cohorts | |||||||||||||
| 11 | MARTHA | >0.05 in >5% samples | Excluded | Included | 388,120 | Sample PCA | Yes | Yes | SWAN | Measured | Array, array position | No | |
| 12 | F5L | >0.05 in >5% samples | Excluded | Excluded | 378,594 | Sample PCA | Yes | Yes | SWAN | Imputed | Array, array position | Family structure | |
Statistical analysis
Two models were run independently by each participating study. Firstly, an additive model for MTHFR 677C>T alone was used to investigate its independent association with genome-wide DNA methylation in a linear manner. For the MTHFR 677C>T variant, genotypes were coded as CC = 0, CT = 1 and TT = 2 to study the effect in methylation per MTHFR 677T allele.
In the second analysis, a weighted Genetic Risk Scores (GRS) was constructed from all the 18 Hcy-associated variants to investigate their combined and additive effect on genome-wide DNA methylation. Weighted GRS were calculated on the basis of their effect sizes [5] and number of corresponding risk alleles. The product of the two was calculated for each SNP and then summed up for all SNPs. The GRS was calculated using the equation below, where N is the number of elevated Hcy causing risk alleles for each SNP (0, 1 or 2 per genotype).
| (1) |
Both the analyses were based on linear mixed models of lme4 package in R. We also analyzed the effect of MTHFR 677C>T and GRS on global methylation levels, where we calculated the overall mean levels per individual as well as categorized the means as per CGI annotations [15]. The models were adjusted for technical covariates and biological covariates like age, sex and differential white blood cell (WBC) counts (see S1 Table for details about covariates for each cohort). The technical covariates were cohort-specific and treated as random effects. WBC counts were either used as measured counts, or they were imputed based on the Houseman method as implemented in the minfi package [17], or the modified version of the Houseman method (Documentation and R script: https://github.com/mvaniterson/wbccPredictor) that uses partial least-squares [18] to handle multivariate responses and high-dimensional covariates and has been previously used [19]. This method from van Iterson used the R package pls [18] to fit the linear model based on the DNA methylation data, to predict the white blood cell composition as percentages that sum up to almost 100%. Age and gender were used as covariates.
Meta-analysis
Summary statistics for the two models were obtained from each study. Because of the different probe exclusions in each cohort [Table 1], we removed probes that were present in ≤4 studies. We also excluded probes with SNPs at single base extension site, and probes with improper binding [12], leaving a total of 465,694 probes for the meta-analysis. Meta-analysis was performed using the fixed effect model in METAL [20], with the classical approach that uses effect size estimates and standard errors as input obtained from the individual study summary statistics for each CpG probe. The output of meta-analysis gave the combined effect size estimates, standard errors and p-values per probe. These p-values were corrected using the Benjamini Hochberg method of false discovery rate (FDR), where FDR <0.05 was considered statistically significant. For the MTHFR 677C>T model, positive effect sizes correspond to percentage increase in methylation per MTHFR 677T allele. For the GRS model, positive effect sizes correspond to percentage increase in methylation per unit increase in GRS. We also took into account heterogeneity of the meta-analysis by I2 which was calculated using METAL per probe and excluded significant probes if I2<40. We also calculated the genomic inflation factor (λ) [21] to estimate the inflation in test statistics that may be caused by population structure or other unknown confounding factors. This λ was estimated for the distribution of p-values using the median method, which is defined as the ratio of the observed median of the test statistic distribution and the expected median 0.455 [21, 22].
Genomic Regions Enrichment of Annotations Tool (GREAT) was used for annotating CpGs for nearby genes, that assigns a basal regulatory region to extend up to 5 kb upstream and 1 kb downstream from its transcription start site and a maximum extension distance up to 1 Mb [23], as defined by UCSC [24]. Furthermore, strength of the instrument or allele score was calculated using the F-statistics, using the tool, mRnd [25]. We took into account cohort heterogeneity I2 and excluded significant probes if I2<40.
Identifying cis- and trans-CpG effects
We defined the CpGs as “cis” when the CpG was annotated within 1Mb upstream or downstream of the SNP. Trans-CpGs were defined as CpGs that were associated with the SNP, and were annotated >1Mb apart. We defined the CpGs in the GRS model the same way by accounting for the bp distance of the CpGs from each of the 18 SNPs. For the significant trans-CpGs that were 1-5Mb apart from any of the 18 SNPs, we performed a conditional analysis adjusting for that SNP to investigate whether they were trans-CpGs associated with Hcy GRS or long range cis-CpGs driven by the nearby SNPs that were part of the GRS. In the conditional analysis, if the bonferroni corrected p-values were no longer significant, we considered those trans-CpGs as long-range cis-CpGs. For the significant trans-CpGs that were >5Mb apart from any of the 18 SNPs, we looked up for their tested individual association with each of the 18 SNPs in our previous trans-CpG mapping analysis [12]. This is to see whether the association of these trans-CpGs was Hcy GRS driven or driven by a single SNP that was a part of GRS. In order to confirm these trans-CpG effects, we performed a similar conditional analysis by including the respective SNP as a covariate in the model. Both conditional analyses were performed on a subset of 3,786 samples from 6 cohorts, and the results were compared with the unconditional analysis in the same subset.
H19/IGF locus
Three Differentially Methylated Regions (DMRs) of IGF2/H19 locus at chromosome 11 have been reported to be related with homocysteine [26, 27]. We identified seven CpGs on the 450k array that were underlying the three DMRs of this locus, for their association with MTHFR 677C>T variant or GRS. Bonferroni method was applied on these 7 CpGs to check for multiple testing.
Enrichment of folate-associated CpGs
We further focused our analysis on the 443 previously identified CPGs, of which methylation in cord blood of newborns were associated with maternal plasma folate levels [28]. We compared the p-values of these 443 CpGs from the MTHFR 677>T and the GRS results, and compared them to the p-values of 100 random CpGs with 1000 permutations, to check for their significant enrichment, using the Fisher’s exact test.
Results
Population characteristics
The meta-analysis included 9,894 adults from 12 cohorts. Studies were population-based, except for the Cohort on Diabetes and Atherosclerosis Maastricht, MARseille THrombosis Association study and the French-Canadian family study, where individuals were selected based on mildly increased diabetes mellitus type 2 and cardiovascular risk factors, cases of venous thrombosis and probands with venous thromboembolism, respectively.
Meta-analysis of MTHFR 677C>T and GRS model
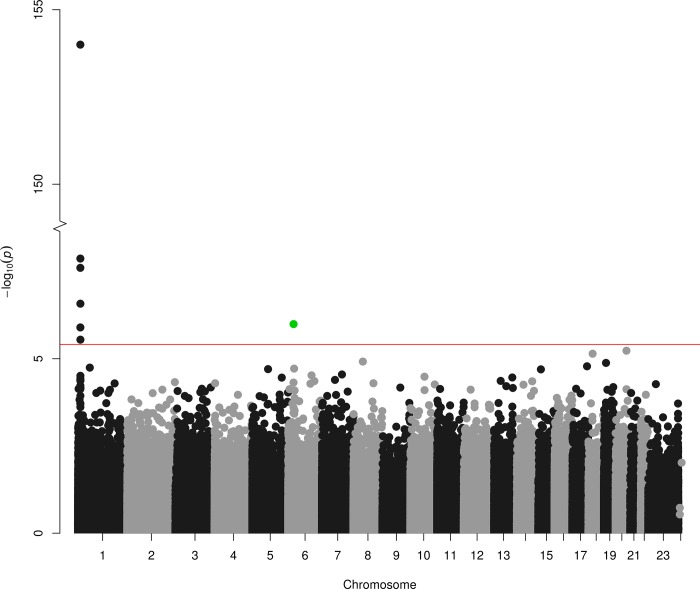
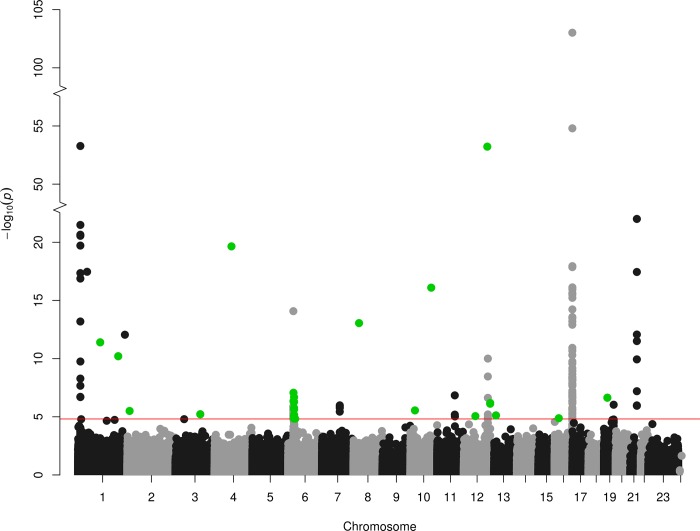
The explained variance in Hcy by MTHFR 677C>T is 5.3% [4] and by GRS is 5.9% [5]. For a sample size of 9,894, the F-statistics of the additive MTHFR 677C>T and GRS was 554 and 621, respectively. Meta-analysis of 456,694 probes identified 35 cis- [S3 Table] and 1 trans- [S5 Table, Table 2] CpGs for the MTHFR model [Fig 1] and 113 cis- [S4 Table] and 30 trans- [Table a in S6 Table, Table a in S7 Table and S8 Table] CpGs for the GRS model [Fig 2]. The λ was 1.01 for the MTHFR 677C>T SNP and 0.92 for GRS [S1 Fig].
Table 2. Genome-wide trans-CpGs with FDR<0.05; associated with the MTHFR 677C>T model or Genetic Risk Score of 18 Hcy-associated variants.
| CpG | N | Beta | SE | P | FDR | I2 | Beta | SE | P | Nearest Genes | Chr | Bp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTHFR 677C>T model | LOOKUP in GRS model | |||||||||||
| cg05411165 | 9894 | -0.005 | 0.001 | 1.02E-06 | 1.40E-02 | 0 | -0.0095 | 0.0038 | 1.16E-02 | ZNF184 (-25414), HIST1H2BL (+309398) | 6 | 27466334 |
| GRS model | LOOKUP in MTHFR 677C>T model | |||||||||||
| cg12805629 | 6277 | -0.018 | 0.004 | 3.15E-06 | 1.23E-02 | 0 | -0.0023 | 0.0011 | 3.81E-02 | MRPL55 (+6698), ARF1 (+19954) | 2 | 11565653 |
| cg08586216+ | 9894 | -0.002 | 0.001 | 1.49E-05 | 4.89E-02 | 37.6 | -0.0003 | 0.0001 | 4.17E-02 | TULP1 (-131681), FKBP5 (+44391) | 6 | 35612351 |
| cg00620062 | 9894 | 0.004 | 0.001 | 2.83E-06 | 1.12E-02 | 0 | 0.0007 | 0.0003 | 3.36E-03 | PTF1A (+6292) | 10 | 23487775 |
| cg00677455* | 9334 | -0.003 | 0.001 | 8.76E-06 | 3.09E-02 | 0 | -0.0004 | 0.0002 | 3.63E-02 | CTDSP2 (-269) | 12 | 58241039 |
| cg01259782 | 6194 | 0.015 | 0.004 | 1.33E-05 | 4.52E-02 | 0 | 0.0021 | 0.0010 | 2.68E-02 | CRYM (+454) | 16 | 21313973 |
Beta: Regression coefficients
SE: Standard errors of the regression coefficients
FDR: False discovery rate adjusted P-value, threshold = 0.05
I2: Heterogeneity I2 parameter
*Promoter-associated
+Enhancer-associated, Enhancer annotation from Illumina 450k annotation
Fig 1. Manhattan plot.
Association between MTHFR 677C>T (rs1801133) and genome-wide DNA methylation in 9,894 samples, with 35 cis-meQTLs at chromosome 1 (black/grey) and 1 trans-meQTL at chromosome 6 (green) with FDR<0.05.
Fig 2. Manhattan plot.
Association between GRS of 18 Hcy-associated SNPs and genome-wide DNA methylation in 9,894 samples, with 113 cis-meQTLs (black/grey) and 30 trans-meQTLs (green), at FDR<0.05.
Cis-CpGs
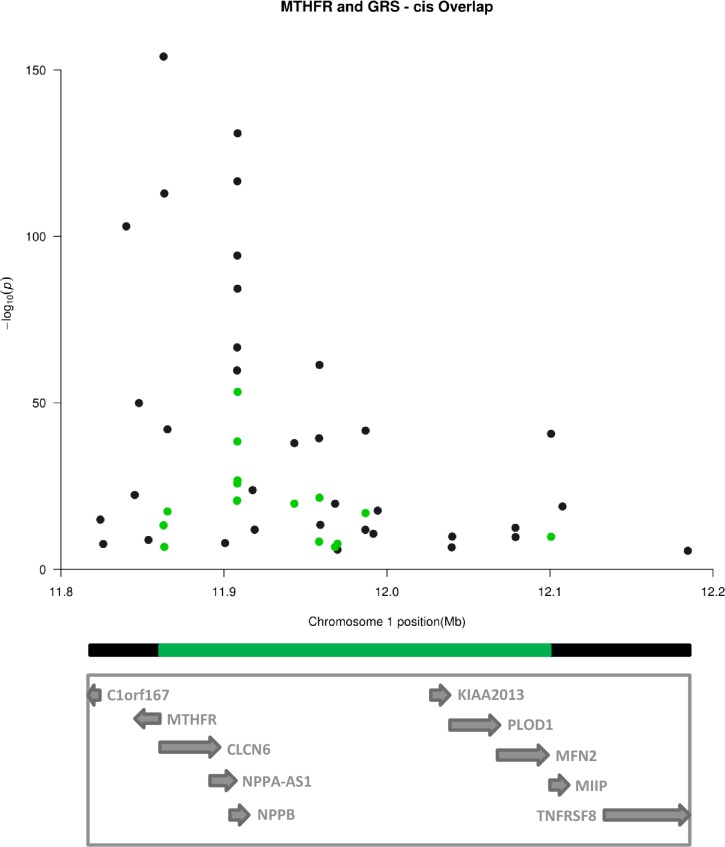
Meta-analysis on 465,694 CpGs of the MTHFR 677C>T variant showed association with 35 cis- CpGs on chromosome 1 with FDR<0.05 [Fig 1, S3 Table]. These cis-CpGs showed a range from 2.4% increase to 1.7% decrease in methylation per MTHFR 677T allele. The nearest genes associated with this cis-region included MTHFR itself, AGTRAP, CLCN6, NPPA, NPPB, PLOD1, MFN2 and TNFRSF8. For the GRS model, we observed 113 cis-CpGs with FDR<0.05 [Fig 2, S4 Table]. Out of the 113, 16 cis-CpGs showed overlap with the MTHFR 677C>T analysis, which involved a smaller region of 238 Kb [Fig 3].
Fig 3. Regional manhattan plot (chr1: 11824095–12184574).
35 (black) and 16 (green) cis-meQTLs of the MTHFR 677C>T and GRS model respectively, in 9,894 samples. The overlap involved a small region of 238 kb (green rectangular line).
Trans-CpGs
For the MTHFR 677C>T model, meta-analysis of 465,694 CpGs identified 1 significant trans-CpG which was located on chromosome 6 [Fig 1, S5 Table]. This trans-CpG (cg05411165) showed 1% decrease in methylation per MTHFR T allele. It was annotated near ZNF184 (25414 bp upstream) and HIST1H2BL (309398 bp downstream). For the GRS model, we observed 30 significant trans-CpGs [Fig 2]. These trans-CpGs showed a range from 5.6% increase to 5.1% decrease in methylation per 0.1 unit increase in GRS. Of these 30 trans-CpGs, 23 were negatively associated with the GRS model. To assess overlap between two models, we evaluated association of the trans-CpG of the MTHFR 677 C>T model within the GRS model. This trans-CpG (cg05411165) showed a 10% decrease in methylation in the GRS model but was not FDR significant (raw p-value = 0.01).
Critical evaluation of the 30 trans-CpGs of GRS model demonstrated that 14 trans-CpGs were located in a large region of 3,08 Mb length within chromosome 6. The GRS model consists of 18 SNPs including a SNP on chromosome 6. The 14 trans CpG identified with the GRS model were at a distance between 1 and 5 Mb away from the Hcy-associated variant rs548987 of the SLC17A3 gene at chromosome 6 [Table a in S6 Table].
Conditional analysis: Chromosome 6 region near rs548987
To investigate whether the 14 trans-CpGs on chromosome 6 near rs548987 were influenced by this variant, we performed conditional analysis on a subset of 3,786 samples from 6 cohorts. After correction of the model for rs548987 as a covariate, none of the 14 trans-CpGs were significant at a bonferroni threshold of 3.57E-03 [Table b in S6 Table].
Conditional analysis: Influence of SNPs within the GRS model
To further investigate the remaining 16 trans-CpGs from the 30, whether any of them were driven by a single variant, rather than the combined effect of the 18 homocysteine-associated variants, we checked the trans-CpG mapping analysis of the single SNPs using the BIOS dataset [12]. We observed that 7 of the 16 remaining trans-CpGs located >5 Mb from the Hcy-associated variants, were directly associated with either rs548987 SNP of SLC17A3 gene at chromosome 6, or rs154657 SNP of DPEP1 gene at chromosome 16 [Table a in S7 Table]. After correction for these 7 trans-CpGs by including the respective SNP as a covariate in the model, none of the 7 trans-CpGs remained significant at a bonferroni threshold of 7.14E-03 [Table b in S7 Table]. After correction for cis-effects of Hcy-associated SNPs in the GRS model, we identified a remaining list of 9 Hcy-associated trans-CpGs, 4 of which had substantial heterogeneity I2 [S8 Table].
Overlapping trans-CpG between MTHFR and GRS models
When doing a lookup in the MTHFR 677C>T model for the finally identified 5 trans-CpGs, all of them showed similar direction of effect, but did not achieve genome-wide significance (Lowest raw p-value = 3.36E-03) [Table 2].
Trans-CpGs affecting Gene expression
We evaluated whether methylation levels of the observed trans-CpG from the MTHFR 677C>T model and 5 trans-CpGs from the GRS model were associated with expression levels of the nearby genes, in the BIOS dataset [12]. None of the trans-CpGs was associated with mRNA expression differences of nearby genes.
H19/IGF2 locus
We specifically focused on the IGF2-H19 region for differential methylation, since methylation at this locus has repeatedly been linked to the homocysteine metabolism in a number of studies [29–31]. S4 Fig shows the results of the whole IGF2-H19 region, and the 7 CpGs annotated to 3 DMRs of the IGF2/H19 gene that had previously been reported to be differentially methylated (DMR0, DMR2, H19-DMR3). Data from our 450k arrays contained 2 CpGs at DMR0, 4 CpGs at DMR2 and 1 CpG at H19-DMR3. None of them showed an association with MTHFR 677C>T or GRS with a Bonferroni cut off of 7.14E-03 [S9 Table, S4 Fig].
Enrichment of previously found folate-associated CpGs
Next we focused on a set of 443 CpGs that were identified to be differentially methylated in children at birth according to the folate levels in the mothers [28]. We found a highly significant enrichment for significant p-values in the MTHFR model in the 443 CpGs as compared to a random set of other CpGs (>3 times enrichment of significant p-values, enrichment p = 0.0079). However, we did not find a significant enrichment for the GRS model.
Global DNA methylation changes
In addition to genome-wide DNA methylation changes we analyzed the effect of MTHFR 677C>T and GRS models on overall mean methylation levels. There was no significant association between the MTHFR 677C>T or GRS on global methylation overall or mean methylation of CpG islands, shores, shelves or non-coding regions [Table 3] [15].
Table 3. Association of MTHFR 677C>T and Genetic Risk Score on mean global methylation levels.
| Methylation | N | I2 | Beta | SE | P |
|---|---|---|---|---|---|
| MTHFR 677C>T | |||||
| GLOBAL | 3,786 | 0.14 | 4.00E-06 | 1.50E-05 | 0.81 |
| CGI | 3,786 | 0.58 | -1.90E-05 | 4.90E-05 | 0.70 |
| SHE | 3,786 | 0.59 | 3.70E-05 | 5.50E-05 | 0.50 |
| SHO | 3,786 | 0.14 | -9.00E-06 | 4.80E-05 | 0.85 |
| NC | 3,786 | 0.61 | 2.70E-05 | 4.90E-05 | 0.57 |
| GRS | |||||
| GLOBAL | 3,786 | 0.67 | -1.30E-05 | 5.60E-05 | 0.81 |
| CGI | 3,786 | 0.07 | -1.84E-04 | 1.81E-04 | 0.31 |
| SHE | 3,786 | 0.00 | 2.29E-04 | 2.01E-04 | 0.25 |
| SHO | 3,786 | 0.52 | -1.20E-04 | 1.76E-04 | 0.50 |
| NC | 3,786 | 0.07 | 1.94E-04 | 1.78E-04 | 0.28 |
Beta: Regression coefficients
SE: Standard errors of the regression coefficients
I2: Heterogeneity I2 parameter
CGI = CpG Islands, SHE = CpG Shelves, SHO = CpG Shores, NC = CpGs at Non-Coding regions
Discussion
This is the first large-scale study to investigate the effect of Hcy-associated SNPs on genome-wide DNA methylation using the Illumina 450k arrays in 9,894 individuals. The results showed no widespread trans-effects of the MTHFR 677C>T SNP on DNA methylation, apart from 1 trans-CpG at chromosome 6. The GRS model showed 5 trans-CpGs, after carefully examining the direct effects of individual SNPs with conditional analyses.
In this current study, we used MTHFR 677C>T independently and the combined weighted GRS of the 18 Hcy-associated variants [5], to test whether mildly elevated Hcy concentrations induce DNA methylation changes in blood cells. Our goal was to investigate genetically defined elevated Hcy on genome-wide DNA methylation. The use of genetic variants is less sensitive to confounding and bias as compared to classical epidemiological studies [32].
We calculated the strength of our exposure variables: MTHFR 677C>T and GRS using the F-statistics. For a strong exposure, the value of the F-statistics is expected to be greater than 10 [33]. With our large sample size (n = 9,894) and proportion of variance explained being 5.3 to 5.9%, the F-statistics was 554 and 621 respectively, indicating very high strength and enough power of our analysis. However, we did not observe widespread trans-effects despite of having strong additive MTHFR 677C>T and GRS.
We observed a single trans-CpG for the MTHFR 677C>T variant. This CpG is located near ZNF184 and HIST1H2BL. Both these genes are thought to play a role in transcriptional regulation. For the GRS, we found 30 trans-CpGs associated, 14 of which are spread over a region of 3,08 Mb at chromosome 6. These CpGs were annotated to genes that included ZNF322 and HIST1H2BJ, HLA-J, HLA-A, HLA-G, but also the proximal region of ZFP57 gene, which was previously identified as a folate-sensitive region in a genome-wide methylation study of 23 women [34]. However, when we performed a conditional analysis on these 14 CpGs in this region, by adjusting for the nearby variant rs548987 of the SLC17A3 gene, the effect sizes significantly attenuated and the nominal p-values were no more significant. The results indicate that this region was influenced by the rs548987 SNP of SLC17A3 gene and was not Hcy-associated.
We finally observed 5 trans-CpGs associated to genetically defined Hcy using the GRS, after carefully examining the direct effects of individual SNPs with conditional analyses and discarding CpGs that showed substantial cohort heterogeneity I2. A total of 3 CpGs showed hypomethylation, one of which was annotated to the FKBP5 gene. FKBP5 encodes for the FK506-binding protein 51 (FKBP51) whose expression has recently been shown to decrease DNMT1 activity and thereby decreasing global methylation [35].
Furthermore, when looking at our methylation-expression results [12], none of the 5 trans-CpGs was associated with mRNA expression differences of nearby genes. The possible explanation for these negative findings could be that these CpG sites might have an effect further away on trans-genes. Conversely, it has been shown that the methylation-expression correlation in cis are not best predicted using the CpG position alone, but by using specific chromatin marks [36]. Furthermore, it could also be that these correlations are specific to other tissues, but not in blood.
We observed that the IGF2-H19 locus did not show association with methylation according to genetically defined elevated homocysteine. This is in contrast to the previous findings in mice, where tissue-specific changes in H19 DMR methylation were found in liver, brain and aorta, and increased expression of H19 was found in aorta [26, 27]. Similar to what we found, the H19-DMR3 between CBS deficient patients and controls also did not show a significant difference, in a previous study [29]. Our results show that this imprinted locus is not deregulated by long-term genetically defined mildly elevated homocysteine. However, previously it was reported that MTHFR 677C>T variant shows changes in DNA methylation in peripheral blood mononuclear cells, only through an interaction with folate [7]. Hence, further studies are needed to study the effect of MTHFR 677C>T variant in the presence of blood folate levels.
We did not see widespread methylation changes associated to mildly elevated plasma Hcy concentrations. This result is not in line with a number of earlier reports, which have shown global methylation changes in association with the MTHFR 677C>T variant and Hcy concentrations [3]. Previous two studies on this topic have shown contradictory results. There has been reports that showed a lower circulating global methylation level in individuals with the MTHFR 677TT genotype [6],[7]. However, there are also a few negative studies that showed no relation between the MTHFR 677TT genotype and global methylation levels [8–10]. All these studies had modest sample sizes (upto 300 individuals were studied), and measured methylation on a global level using the LINE-1 assay, which measures a repetitive sequence, of which the function is unknown. In contrast, we here studied a genome wide site-specific analysis focused on functional regions of the genome [15]. We here show convincing evidence that there is no association between the MTHFR 677C>T or GRS on overall methylation levels, nor is there a relationship between methylation of CpG islands, shores, shelves or non-coding regions separately, which supports the previous null associations.
Furthermore, in order to test for causal effect in a mendelian randomization study, an instrument, which is in our case MTHFR 677C>T or GRS, should satisfy the 3 basic assumptions [37, 38]. One, the instrument should be associated with the exposure, which is in our case Hcy. Two, the instrument should not affect the outcome, which is in our case DNA methylation, except through the exposure Hcy. Three, the instrument should not be associated with any confounder of the exposure-outcome association. Although assumptions one and two are satisfied in our case [5], the GRS model might violate assumption three [37, 38].
The GRS model contains a few SNPs which are, in addition to the association with Hcy, also associated with other traits. For example, the variants near to HNF1A gene have been associated with a number of other traits [39–44]. This could also be the reason why the results of the MTHFR 677C>T and GRS models are quite different. Nevertheless, the MTHFR 677C>T variant explains most of the variation in Hcy, as compared to the other variants in the GRS model and is therefore a strong instrument to examine the effect of deregulation of the one-carbon metabolism on methylation.
The relationship between MTHFR 677C>T and DNA methylation is modified by folate levels. Only in individuals with low folate status, the effect of the MTHFR 677TT genotype is seen [7]. Unfortunately, we were unable to study this interaction, since folate levels were not available in our study. Another prerequisite to be able to perform MR is that the relationship between Hcy and methylation is known. The relationship between Hcy and DNA methylation is only known in studies until now where methylation is measured at a global level. Therefore, the estimation of the causal effect could not be done. Unfortunately, we also did not have Hcy data available in all cohorts of this study, and therefore were unable to perform a full mendelian randomization study. We rather focused on the association of genetically defined elevated Hcy levels with DNA methylation.
We did not find widespread differences in methylation related to genetically defined homocysteine levels. The association was not observed in global methylation levels nor in widespread CpGs including the previously known H19/IGF2 locus. There are a number of possible explanations for this finding. First, it is known that the relationship between MTHFR 677C>T and DNA methylation is modified by folate levels, as described above. The effect of MTHFR 677C>T is seen in individuals with low folate status [7], which could have masked possible relationships between the MHTFR variant and methylation. A second possible explanation for the relative low number of identified CpGs, is that we have studied the wrong tissue. Most methylation measures are conducted in blood leukocytes as this tissue is readily available. However, the causal effect of Hcy could be specific to other tissues like liver, heart and brain. Therefore, the possible effect of mildly elevated Hcy on such specific tissues cannot be excluded. Third, there is little variation in the one-carbon metabolism in the normal population. This metabolism is pivotal to cell survival and function and therefore tidily regulated. It could be that there is a correlation between homocysteine and more pronounced effects on methylation when homocysteine levels are more extreme.
Conclusions
We observed 1 trans-CpG (nearest genes ZNF184 and HIST1H2BL) on chromosome 6 associated with the MTHFR 677C>T variant. The GRS model showed 5 significant trans-CpGs, which do not overlap with the MTHFR trans-CpG. In conclusion, our results do not show widespread statistically significant trans-effects of MTHFR and GRS models, and therefore do not support the hypothesis that genetically defined mildly elevated Hcy concentrations are associated with widespread methylation changes in leukocytes. More studies with measured Hcy concentrations are needed to confirm this.
Supporting information
Association of (a) MTHFR 677C>T and (b) Genetic risk score of all the 18 Hcy-associated variants with genome-wide DNA methylation.
(PDF)
Trans-meQTL of the MTHFR 677C>T model across 12 cohorts.
(PDF)
Trans-meQTLs of the Genetic risk score model across 12 cohorts.
(PDF)
(a) MTHFR 677 C>T variant or (b) Genetic risk score associated 3 DMRs of IGF2/H19 genes containing 7 CpGs (green) from the 450k data; DMR0 “Chr.11:2,170,380–2,170,517” with 2 CpGs, DMR2 “Chr.11:2,154,113–2,154,414” with 4 CpGs and H19-DMR3 “Chr.11:2,021,072–2,021,273” with 1 CpG.
(PDF)
(PDF)
Quality control of SNPs and imputation.
(PDF)
The CpGs are sorted in base pairs.
(PDF)
The CpGs are sorted in chromosomes and base pairs.
(PDF)
(PDF)
(a) 14 trans-meQTLs of chromosome 6 with FDR<0.05 that are associated with Genetic Risk Score of 18 Hcy-associated variants in a sample size of 9,894, and were 1Mb-5Mb away from the SNP rs548987 of SLC17A3 gene. (b) Conditional analysis for the 14 trans-meQTLs of chromosome 6 with adjustment for the SNP rs548987 of SLC17A3 gene in a subset of 3,786. The CpGs are sorted in base pairs.
(PDF)
(a) 7 genome-wide trans-meQTLs with FDR<0.05 that are associated with Genetic Risk Score of 18 Hcy-associated variants in a sample size of 9,894 and were a direct trans-meQTL of either SNP rs548987 of SLC17A3 gene or rs154657 SNP of DPEP1 gene. (b) Conditional analysis for the 7 genome-wide trans-meQTLs with adjustment for their respective SNP rs548987 of SLC17A3 gene at chromosome 6 or rs154657 SNP of DPEP1 gene at chromosomes 16 in a subset of 3,786.
(PDF)
The CpGs are sorted in chromosomes and base pairs.
(PDF)
(a) MTHFR 677C>T variant associated DMPs at the 3 IGF2/H19 DMR regions at chromosome 11. (b) GRS associated DMPs at the 3 IGF2/H19 DMR regions at chromosome 11.
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
Rotterdam Study
The generation and management of the Illumina 450K methylation array data (EWAS data) for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We thank Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins and Mr. Marijn Verkerk for their help in creating the methylation database.
The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Biobank-Based Integrative Omics Studies (BIOS) Consortium
Samples were contributed by LifeLines (http://lifelines.nl/lifelines-research/general), the Leiden Longevity Study (http://www.leidenlangleven.nl), the Netherlands Twin Registry (http://www.tweelingenregister.org), the Rotterdam studies (http://www.erasmus-epidemiology.nl/research/ergo.htm), the CODAM study (http://www.carimmaastricht.nl/), and the PAN study (http://www.alsonderzoek.nl/). We thank the participants of all aforementioned biobanks and acknowledge the contributions of the investigators to this study, especially Aaron Isaacs, René Pool, Marian Beekman, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Jan Bot, Patrick Deelen, Irene Nooren, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, and Morris A. Swertz (Additional file 1). This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative.
We acknowledge the support from the Netherlands CardioVascular Research Initiative (the Dutch Heart Foundation, Dutch Federation of University Medical Centres, the Netherlands Organisation for Health Research and Development, and the Royal Netherlands Academy of Sciences) for the GENIUS project “Generating the best evidence-based pharmaceutical targets for atherosclerosis” (CVON2011-19). This work was performed within the framework of the Biobank-Based Integrative Omics Studies (BIOS) Consortium funded by BBMRI-NL, a research infrastructure financed by the Dutch government (NWO 184.021.007).
Data Availability
We are not allowed to publish the full data on internet because of privacy policy. However, data can be accessed by individual researchers through these contacts for each individual study: Requests for the data from the Rotterdam study may be sent to: Frank van Rooij (f.vanrooij@erasmusmc.nl). Requests for the data from the Lothian Birth Cohort may be sent to Riccardo Marioni (rmarioni@exseed.ed.ac.uk). LBC methylation data have been submitted to the European Genome-phenome Archive -accession number EGAS00001000910. The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA-gen (https://epi.helmholtz-muenchen.de/). Data requests can be submitted online and are subject to approval by the KORA Board. Requests for the data from the Framingham Heart Study may be sent to Roby Joehanes (robyjoehanes@hsl.harvard.edu) or from the link: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000724.v6.p10. Requests for the data from the Biobank-Based Integrative Omics Studies (BIOS) Consortium (RS, LLS, LL, CODAM and NTR cohorts) may be sent to Rick Jansen (Ri.Jansen@ggzingeest.nl) and from the accession number EGAC00001000277. Requests for the data from the MARTHA study may be sent to David-Alexandre Trégouët (david.tregouet@upmc.fr). MARTHA Methylation data are already available in the Array Express database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3127. Request for the data from the F5L study may be sent to France Gagnon (france.gagnon@utoronto.ca).
Funding Statement
Rotterdam Study: The generation and management of the Illumina 450K methylation array data (EWAS data) for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The EWAS data was funded by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by the Netherlands Organization for Scientific Research (NWO; project number 184021007) and made available as a Rainbow Project (RP3; BIOS) of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL). We thank Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins, Mr. Marijn Verkerk, and Lisette Stolk PhD for their help in creating the methylation database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. Biobank-Based Integrative Omics Studies (BIOS) Consortium Samples were contributed by LifeLines (http://lifelines.nl/lifelines-research/general), the Leiden Longevity Study (http://www.leidenlangleven.nl), the Netherlands Twin Registry (http://www.tweelingenregister.org), the Rotterdam studies (http://www.erasmus-epidemiology.nl/research/ergo.htm), the CODAM study (http://www.carimmaastricht.nl/), and the PAN study (http://www.alsonderzoek.nl/). We thank the participants of all aforementioned biobanks and acknowledge the contributions of the investigators to this study, especially Aaron Isaacs, René Pool, Marian Beekman, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Jan Bot, Patrick Deelen, Irene Nooren, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, and Morris A. Swertz (S1_File-BIOSconsortiumMembershipList). This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative. We acknowledge the support from the Netherlands CardioVascular Research Initiative (the Dutch Heart Foundation, Dutch Federation of University Medical Centres, the Netherlands Organisation for Health Research and Development, and the Royal Netherlands Academy of Sciences) for the GENIUS project “Generating the best evidence-based pharmaceutical targets for atherosclerosis” (CVON2011-19). This work was performed within the framework of the Biobank-Based Integrative Omics Studies (BIOS) Consortium funded by BBMRI-NL, a research infrastructure financed by the Dutch government (NWO 184.021.007). MARseille THrombosis Association Study (MARTHA) D.A. was supported by a PhD grant from the Région Ile de France (CORDDIM). Statistical analyses of the MARTHA were performed using the C2BIG computing cluster, funded by the Région Ile de France, Pierre and Marie Curie University, and the ICAN Institute for Cardiometabolism and Nutrition (ANR-10-IAHU-05). Framingham Heart Study (FHS): The Framingham Heart Study is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute Bethesda, MD. Cooperative health research in the Region of Augsburg (KORA) The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research has been supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The research leading to these results has received funding fromthe European Union Seventh Framework Programme under grant agreement [n°603288] (Systems Biology to Identify Molecular Targets for Vascular Disease Treatment – SysVasc; http://www.sysvasc.eu/). Lothian Birth Cohort (LBC): Phenotype collection in the Lothian Birth Cohort 1921 was supported by the UK’s Biotechnology and Biological Sciences Research Council (BBSRC), The Royal Society and The Chief Scientist Office of the Scottish Government. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Age UK (The Disconnected Mind project). Methylation typing was supported by the Centre for Cognitive Ageing and Cognitive Epidemiology (Pilot Fund award), Age UK, The Wellcome Trust Institutional Strategic Support Fund, The University of Edinburgh, and The University of Queensland. REM, JMS, and IJD are members of the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology (CCACE). CCACE is supported by funding from the BBSRC, the Medical Research Council (MRC), and the University of Edinburgh as part of the cross-council Lifelong Health and Wellbeing initiative (MR/K026992/1). French-Canadian Family Study on Factor V Leiden (F5L) Thrombophilia (F5L Family Study) The F5L Family Study was supported by the Canadian Institutes of Health Research (Grant MOP 86466) and by the Heart and Stroke Foundation of Canada (Grant T6484). F. Gagnon is recipient of a Canada Research Chair.
References
- 1.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002;21:5400–13. doi: 10.1038/sj.onc.1205651 [DOI] [PubMed] [Google Scholar]
- 2.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA Methylation in Cancer and Aging. Cancer Res 2016;76:3446–50. doi: 10.1158/0008-5472.CAN-15-3278 [DOI] [PubMed] [Google Scholar]
- 3.Mandaviya PR, Stolk L, Heil SG. Homocysteine and DNA methylation: a review of animal and human literature. Mol Genet Metab 2014;113:243–52. doi: 10.1016/j.ymgme.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 4.Borges MC, Hartwig FP, Oliveira IO, Horta BL. Is there a causal role for homocysteine concentration in blood pressure? A Mendelian randomization study. Am J Clin Nutr 2016;103:39–49. doi: 10.3945/ajcn.115.116038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, et al. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr 2013;98:668–76. doi: 10.3945/ajcn.112.044545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro R, Rivera I, Ravasco P, Camilo ME, Jakobs C, Blom HJ, et al. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C—>T and 1298A—>C mutations are associated with DNA hypomethylation. J Med Genet 2004;41:454–8. doi: 10.1136/jmg.2003.017244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A 2002;99:5606–11. doi: 10.1073/pnas.062066299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayanan S, McConnell J, Little J, Sharp L, Piyathilake CJ, Powers H, et al. Associations between two common variants C677T and A1298C in the methylenetetrahydrofolate reductase gene and measures of folate metabolism and DNA stability (strand breaks, misincorporated uracil, and DNA methylation status) in human lymphocytes in vivo. Cancer Epidemiol Biomarkers Prev 2004;13:1436–43. [PubMed] [Google Scholar]
- 9.Hanks J, Ayed I, Kukreja N, Rogers C, Harris J, Gheorghiu A, et al. The association between MTHFR 677C>T genotype and folate status and genomic and gene-specific DNA methylation in the colon of individuals without colorectal neoplasia. Am J Clin Nutr 2013;98:1564–74. doi: 10.3945/ajcn.113.061432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pufulete M, Al-Ghnaniem R, Rennie JA, Appleby P, Harris N, Gout S, et al. Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br J Cancer 2005;92:838–42. doi: 10.1038/sj.bjc.6602439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. bioRxiv 2015. [DOI] [PubMed]
- 13.Pidsley R, CC YW, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 2013;14:293 doi: 10.1186/1471-2164-14-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol 2012;13:R44 doi: 10.1186/gb-2012-13-6-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slieker RC, Bos SD, Goeman JJ, Bovee JV, Talens RP, van der Breggen R, et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013;6:26 doi: 10.1186/1756-8935-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonder MJ, Kasela S, Kals M, Tamm R, Lokk K, Barragan I, et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics 2014;15:860 doi: 10.1186/1471-2164-15-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. doi: 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mevik B.H, Wehrens R. The pls Package: Principal Component and Partial Least Squares Regression in R. Journal of Statistical Software 2007;18. [Google Scholar]
- 19.Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol 2016;17:138 doi: 10.1186/s13059-016-1000-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999;55:997–1004. [DOI] [PubMed] [Google Scholar]
- 22.Tsepilov YA, Ried JS, Strauch K, Grallert H, van Duijn CM, Axenovich TI, et al. Development and application of genomic control methods for genome-wide association studies using non-additive models. PLoS One 2013;8:e81431 doi: 10.1371/journal.pone.0081431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. doi: 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res 2016;44:D717–25. doi: 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 2013;42:1497–501. doi: 10.1093/ije/dyt179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devlin AM, Bottiglieri T, Domann FE, Lentz SR. Tissue-specific changes in H19 methylation and expression in mice with hyperhomocysteinemia. J Biol Chem 2005;280:25506–11. doi: 10.1074/jbc.M504815200 [DOI] [PubMed] [Google Scholar]
- 27.Glier MB, Ngai YF, Sulistyoningrum DC, Aleliunas RE, Bottiglieri T, Devlin AM. Tissue-specific relationship of S-adenosylhomocysteine with allele-specific H19/Igf2 methylation and imprinting in mice with hyperhomocysteinemia. Epigenetics 2013;8:44–53. doi: 10.4161/epi.23063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 2016;7:10577 doi: 10.1038/ncomms10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heil SG, Riksen NP, Boers GH, Smulders Y, Blom HJ. DNA methylation status is not impaired in treated cystathionine beta-synthase (CBS) deficient patients. Mol Genet Metab 2007;91:55–60. doi: 10.1016/j.ymgme.2007.01.008 [DOI] [PubMed] [Google Scholar]
- 30.Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, Jones A, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med 2011;28:481–7. doi: 10.3892/ijmm.2011.754 [DOI] [PubMed] [Google Scholar]
- 31.Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, et al. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol 2008;320:79–91. doi: 10.1016/j.ydbio.2008.04.025 [DOI] [PubMed] [Google Scholar]
- 32.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res 2012;21:223–42. doi: 10.1177/0962280210394459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 2011;40:740–52. doi: 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, et al. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. Faseb J 2014;28:4068–76. doi: 10.1096/fj.13-249029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gassen NC, Fries GR, Zannas AS, Hartmann J, Zschocke J, Hafner K, et al. Chaperoning epigenetics: FKBP51 decreases the activity of DNMT1 and mediates epigenetic effects of the antidepressant paroxetine. Sci Signal 2015;8:ra119 doi: 10.1126/scisignal.aac7695 [DOI] [PubMed] [Google Scholar]
- 36.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol 2014;15:R37 doi: 10.1186/gb-2014-15-2-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson SA, Hernan MA. Commentary: how to report instrumental variable analyses (suggestions welcome). Epidemiology 2013;24:370–4. doi: 10.1097/EDE.0b013e31828d0590 [DOI] [PubMed] [Google Scholar]
- 38.Martens EP, Pestman WR, de Boer A, Belitser SV, Klungel OH. Instrumental variables: application and limitations. Epidemiology 2006;17:260–7. doi: 10.1097/01.ede.0000215160.88317.cb [DOI] [PubMed] [Google Scholar]
- 39.Ligthart S, de Vries PS, Uitterlinden AG, Hofman A, group CIw, Franco OH, et al. Pleiotropy among common genetic loci identified for cardiometabolic disorders and C-reactive protein. PLoS One 2015;10:e0118859 doi: 10.1371/journal.pone.0118859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Below JE, Gamazon ER, Morrison JV, Konkashbaev A, Pluzhnikov A, McKeigue PM, et al. Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia 2011;54:2047–55. doi: 10.1007/s00125-011-2188-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonnycastle LL, Willer CJ, Conneely KN, Jackson AU, Burrill CP, Watanabe RM, et al. Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns. Diabetes 2006;55:2534–40. doi: 10.2337/db06-0178 [DOI] [PubMed] [Google Scholar]
- 42.Lauc G, Essafi A, Huffman JE, Hayward C, Knezevic A, Kattla JJ, et al. Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet 2010;6:e1001256 doi: 10.1371/journal.pgen.1001256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegele RA, Cao H, Harris SB, Hanley AJ, Zinman B, Connelly PW. The private hepatocyte nuclear factor-1alpha G319S variant is associated with plasma lipoprotein variation in Canadian Oji-Cree. Arterioscler Thromb Vasc Biol 2000;20:217–22. [DOI] [PubMed] [Google Scholar]
- 44.Pierce BL, Ahsan H. Genome-wide "pleiotropy scan" identifies HNF1A region as a novel pancreatic cancer susceptibility locus. Cancer Res 2011;71:4352–8. doi: 10.1158/0008-5472.CAN-11-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association of (a) MTHFR 677C>T and (b) Genetic risk score of all the 18 Hcy-associated variants with genome-wide DNA methylation.
(PDF)
Trans-meQTL of the MTHFR 677C>T model across 12 cohorts.
(PDF)
Trans-meQTLs of the Genetic risk score model across 12 cohorts.
(PDF)
(a) MTHFR 677 C>T variant or (b) Genetic risk score associated 3 DMRs of IGF2/H19 genes containing 7 CpGs (green) from the 450k data; DMR0 “Chr.11:2,170,380–2,170,517” with 2 CpGs, DMR2 “Chr.11:2,154,113–2,154,414” with 4 CpGs and H19-DMR3 “Chr.11:2,021,072–2,021,273” with 1 CpG.
(PDF)
(PDF)
Quality control of SNPs and imputation.
(PDF)
The CpGs are sorted in base pairs.
(PDF)
The CpGs are sorted in chromosomes and base pairs.
(PDF)
(PDF)
(a) 14 trans-meQTLs of chromosome 6 with FDR<0.05 that are associated with Genetic Risk Score of 18 Hcy-associated variants in a sample size of 9,894, and were 1Mb-5Mb away from the SNP rs548987 of SLC17A3 gene. (b) Conditional analysis for the 14 trans-meQTLs of chromosome 6 with adjustment for the SNP rs548987 of SLC17A3 gene in a subset of 3,786. The CpGs are sorted in base pairs.
(PDF)
(a) 7 genome-wide trans-meQTLs with FDR<0.05 that are associated with Genetic Risk Score of 18 Hcy-associated variants in a sample size of 9,894 and were a direct trans-meQTL of either SNP rs548987 of SLC17A3 gene or rs154657 SNP of DPEP1 gene. (b) Conditional analysis for the 7 genome-wide trans-meQTLs with adjustment for their respective SNP rs548987 of SLC17A3 gene at chromosome 6 or rs154657 SNP of DPEP1 gene at chromosomes 16 in a subset of 3,786.
(PDF)
The CpGs are sorted in chromosomes and base pairs.
(PDF)
(a) MTHFR 677C>T variant associated DMPs at the 3 IGF2/H19 DMR regions at chromosome 11. (b) GRS associated DMPs at the 3 IGF2/H19 DMR regions at chromosome 11.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
We are not allowed to publish the full data on internet because of privacy policy. However, data can be accessed by individual researchers through these contacts for each individual study: Requests for the data from the Rotterdam study may be sent to: Frank van Rooij (f.vanrooij@erasmusmc.nl). Requests for the data from the Lothian Birth Cohort may be sent to Riccardo Marioni (rmarioni@exseed.ed.ac.uk). LBC methylation data have been submitted to the European Genome-phenome Archive -accession number EGAS00001000910. The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA-gen (https://epi.helmholtz-muenchen.de/). Data requests can be submitted online and are subject to approval by the KORA Board. Requests for the data from the Framingham Heart Study may be sent to Roby Joehanes (robyjoehanes@hsl.harvard.edu) or from the link: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000724.v6.p10. Requests for the data from the Biobank-Based Integrative Omics Studies (BIOS) Consortium (RS, LLS, LL, CODAM and NTR cohorts) may be sent to Rick Jansen (Ri.Jansen@ggzingeest.nl) and from the accession number EGAC00001000277. Requests for the data from the MARTHA study may be sent to David-Alexandre Trégouët (david.tregouet@upmc.fr). MARTHA Methylation data are already available in the Array Express database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3127. Request for the data from the F5L study may be sent to France Gagnon (france.gagnon@utoronto.ca).