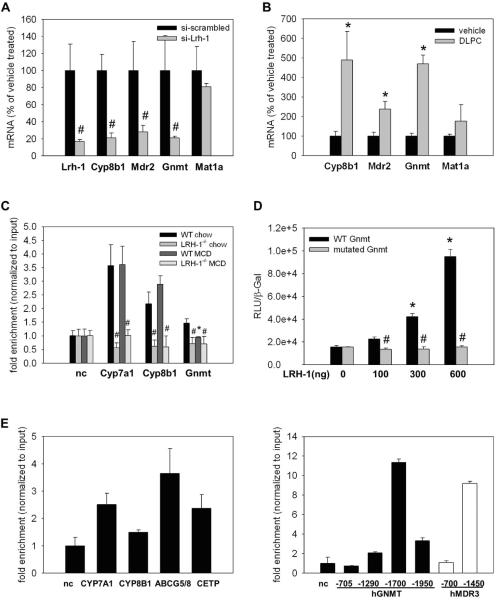
Figure 4. LRH-1 is a transcriptional regulator of key enzymes of the methyl-pool cycle.
A. AML-12 cells were treated with scrambled siRNA or a pool of 3 different Lrh-1 siRNAs for 24h. The classical transcriptional Lrh-1 target Cyp8b1 is significantly reduced in line with reduction of mRNA levels of Mdr2 and Gnmt. B. AML-12 cells were treated with vehicle or the LRH-1 ligand dilauroyl-sn-glycero-3-phosphocholine (DLPC) for 24h. mRNA levels of the classical Lrh-1 target gene Cyp8b1 are significantly increased in line with an induction of Mdr2 and Gnmt mRNA. n=triplicates. * p<0.05, vehicle vs. DLPC; # p<0.05, si-scrambled vs. si-Lrh-1. Error bars represent means ± standard deviation. C. Wildtype (WT) and liver specific LRH-1 knockout (Lrh-1−/−) mice were fed MCD diet for 2 weeks. LRH-1 ChIP-PCR for LRH-1 binding sites in the promoters of Cyp7a1, Cyp8b1 and Gnmt; n=3 mice per group. D. Luciferase assay using the minimal promoter of Gnmt (−661 to TSS) unmutated (WT Gnmt) or after mutation of the ChIPed binding sites at −143 (mutated Gnmt). E. LRH-1 ChIP-PCR from pooled liver tissue from normal control livers biopsies for classical LRH-1 target genes (left panel) and for human GNMT binding sites (right panel, black bars) and MDR3 binding sites (right panel, white bars). nc, negative control.