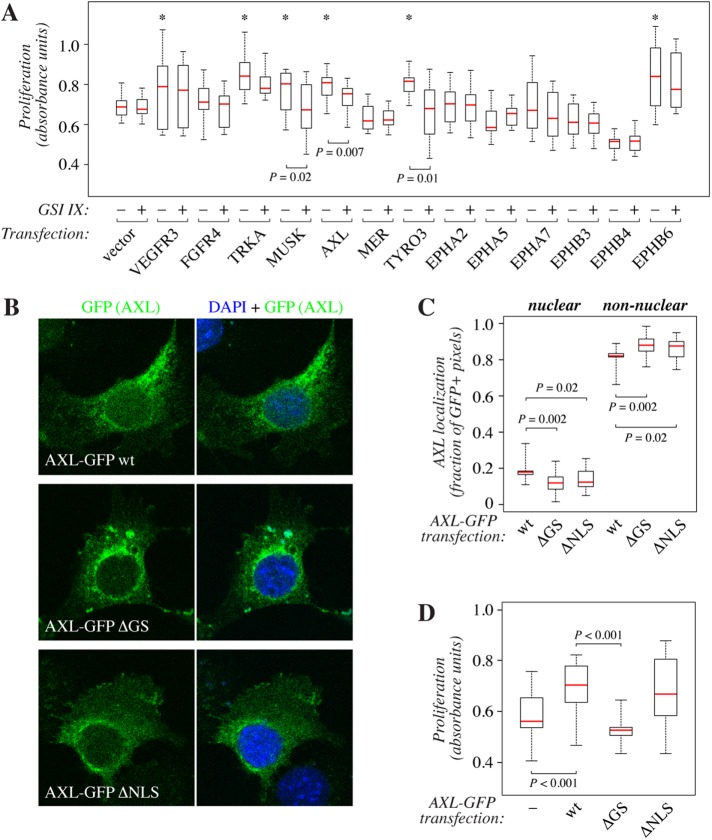
FIGURE 4:
Functional analysis of gamma-secretase–mediated RTK cleavage. (A) WST-8 cell proliferation analysis of NIH-3T3 transfectants expressing novel cleavable RTKs VEGFR3, FGFR4, TRKA, MUSK, AXL, MER, TYRO3, EPHA2, EPHA5, EPHA7, EPHB3, EPHB4, or EPHB6. The cells were treated for 72 h in the presence or absence of 5 μM GSI IX. Data from at least six parallel samples from three independent experiments are shown. An asterisk (*) on top of a box plot indicates significantly (p < 0.05) increased proliferation as compared with the vector control transfectant in the absence of GSI IX. (B) Confocal microscopy analysis of NIH-3T3 cells overexpressing C-terminally GFP-tagged wild-type AXL (wt), AXL with a mutated gamma-secretase cleavage site (∆GS), or AXL with a mutated nuclear localization signal (∆NLS). GFP signal is shown in green, DAPI-stained nuclei in blue. (C) Quantification of the confocal immunofluorescence analysis of AXL-GFP localization shown in B. Nuclear localization is presented as the percentage of GFP-specific signals colocalizing with DAPI of all GFP-specific signals within the cells. At least 20 cells were analyzed per construct. (D) A 72-h WST-8 cell proliferation analysis of NIH-3T3 transfectants expressing wild-type AXL (wt) or AXL ∆GS or AXL ∆NLS mutant constructs.