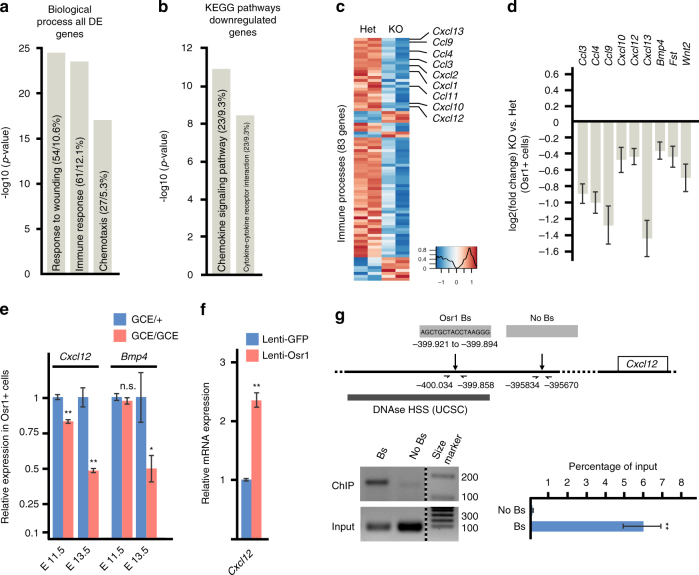
Fig. 8.
Osr1 cells are a source or growth factor signalling and Osr1 directly controls Cxcl12 expression. a Gene ontology (GO) analysis for “biological process” using all deregulated (DE) genes (Osr1 GCE/+ vs. Osr1 GCE/GCE cells) showed significant enrichment for the indicated terms. b KEGG pathway analysis of downregulated genes. c Heatmap depiction of all non-redundant DE genes contained in the GO terms shown in c collectively termed “immune processes”. Several genes for signalling molecules from chemokine families appear downregulated in Osr1 GCE/GCE limb cells. d Deregulation of secreted signalling molecules selected from all DE genes for their previously demonstrated role in myogenesis. e Confirmation of downregulation of Cxcl12 and Bmp4 in Osr1-deficient cells isolated at E11.5 and E13.5 by RT-qPCR (n = 5). f Upregulation of Cxcl12 upon lentiviral Osr1 overexpression. g Chromatin immunoprecipitation (ChIP) for a putative Cxcl12 enhancer. Representative gel pictures of ChIP vs. input are shown below schematic representation of the locus, confirmation of binding site pulldown via RT-qPCR (n = 3) shown right. Bs: putative binding site, No Bs: control region. DNAse HSS: DNAseI hypersensitive regions according to UCSC. Size marker: 100 base pair ladder; band sizes are indicated in base pairs. Error bars represent log2 fold change standard error (ifcSE) estimated by DESeq2 in d and s.e.m. in e, f, g. T-test: *=p < 0.05; **=p < 0.01. N-numbers indicate biological replicates, i.e. samples from different specimen in e or independent assays in f, g