Fig. 9.
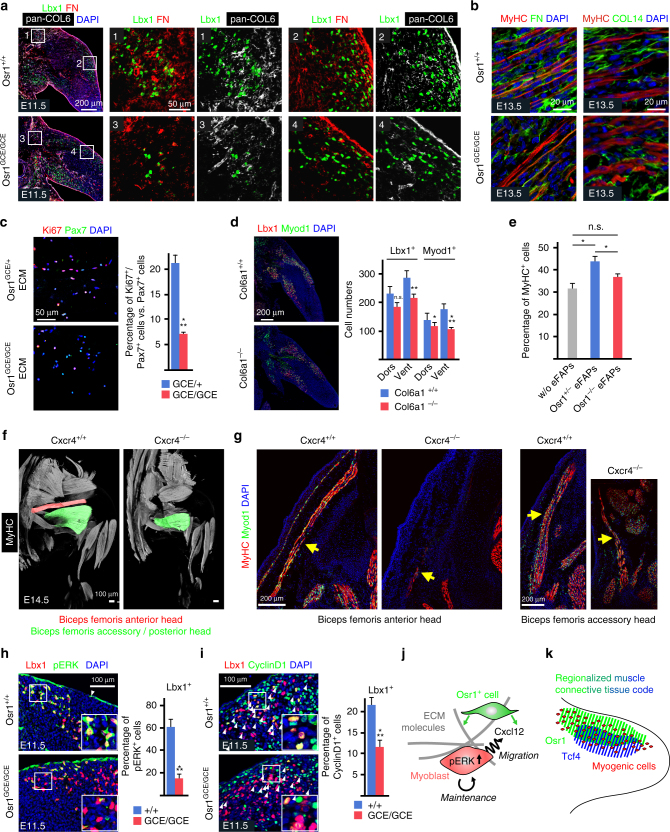
Osr1-deficiency impairs the myogenic niche in developing limbs. a Decreased abundance of COL6 and FN (Fibronectin) in proximal (inserts 1 and 3) and dorsal (inserts 2 and 4) muscle primordia of Osr1-deficient E11.5 limbs. Boxed regions are shown in the numbered panels to the right and are magnifications of the area. b Reduced abundance of interstitial FN and disarrangement of COL14 in E13.5 Osr1 GCE/GCE limb muscles as compared to wild-type littermate controls. c Extracellular matrix (ECM) produced by Osr1GCE/+ cells, but not ECM produced by Osr1GCE/GCE cells supports myoblast proliferation (n = 3). d E11.5 Col6a1 −/− embryos show a decrease in myogenic progenitor numbers (n = 3). e Transwell assay shows a beneficial effect of E13.5 Osr1GCE/+ embryonic FAPs (eFAPs) but not Osr1GCE/GCE eFAPs on myogenesis. Transwell culture without eFAPs was used as control (n = 3). f Cxcr4 −/− embryos show muscle defects overlapping with Osr1GCE/GCE embryos. g Sections of Cxcr4 +/+ and Cxcr4 −/− embryos show almost complete reduction of the biceps femoris anterior head and hypoplasia of the biceps femoris accessory head. h Decreased phosphorylated ERK (pERK) and (i) CyclinD1 in myogenic Lbx1+ cells in E11.5 Osr1 mutants (n = 3). j Schematic model of Osr1+ cell function. Osr1+ cells (green) produce chemokines as Cxcl12 and a specific extracellular matrix (ECM) supporting myoblast proliferation and survival involving pERK signaling. Cxcl12 in addition likely provides local migratory cues ensuring correct spatial distribution of myoblasts as a prerequisite for muscle patterning. k Schematic model of limb muscle connective tissue compartmentalization. Specific transcription factors (Osr1 and Tcf4 are shown as examples) are expressed regionally in the limb bud with areas of overlap creating a regionalized code that likely determines the composition of the local niche. Error bars represent s.e.m. T-test: *=p < 0.05; **=p < 0.01; ***=p < 0.001. N-numbers indicate biological replicates, i.e. samples from different specimen in d, h, i or independent assays in c, e