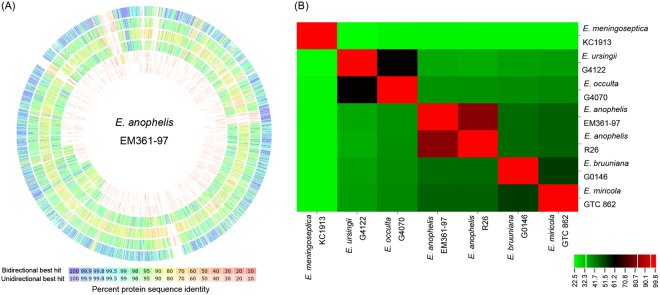
Figure 3.
Genomic comparison among Elizabethkingia species. (A) The genome of E. anophelis EM361-97 (center) compared to E. anophelis R26T (the outermost circle; ring 1), E. bruuniana G0146T (ring 2), E. meningoseptica KC1913T (ring 3), E. miricola GTC 862 T (ring 4), E. occulta G4070T (ring 5), and E. ursingii G4122T (the innermost circle; ring 6). The genome of E. anophelis EM361-97 was highly similar to the type strain of E. anophelis R26T. (B) The in silico DNA-DNA-hybridization (DDH) values between different strains calculated using the Genome-to-Genome Distance Calculator. The DDH value between E. anophelis EM361-97 and E. anophelis R26T was 82%. Elizabethkingia meningoseptica KC1913T demonstrated a relatively large phylogenetic distance from other strains of Elizabethkingia.