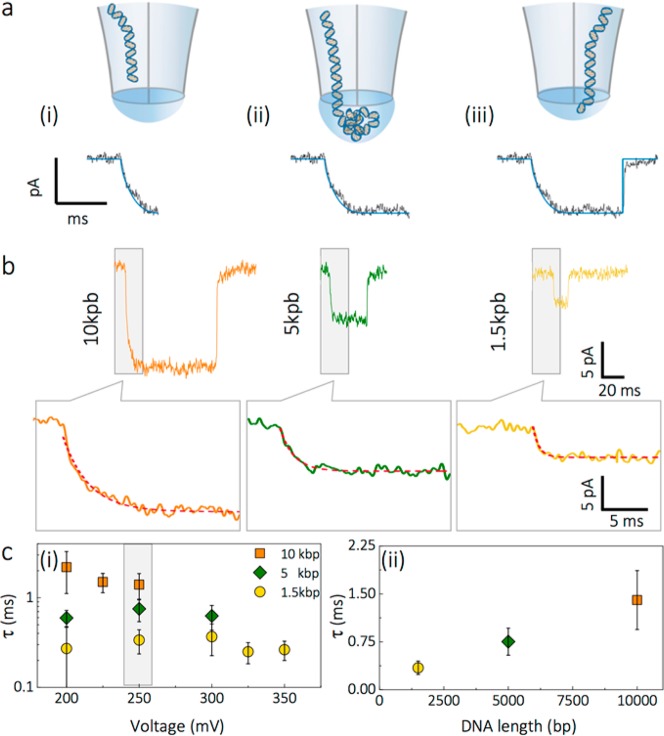
Figure 3.
DNA threading model in nanobridge configuration. (a) Schematic of the threading process: (i) The dsDNA molecule is threaded inside the nanobridge leading to its expansion. The threading process results in the ionic current exhibiting a monoexponential decay with time constant τ. (ii) The DNA recoils inside the bridge until the surface energy of the bridge matches the energy of the DNA confinement. As the DNA in the droplet is predominately governed by Brownian motion, the duration of the blockade is governed by the time it takes the DNA to rearrange and become inserted and finally (iii) threads into the second barrel. (b) Examples of 10, 5, and 1.5 kbp DNA translocation events recorded in nanobridge configuration in 100 mM KCl. The onset of each translocation event was fit with a monoexponential decay function. (c) Dependence of threading time τ on voltage applied (left panel) for 10, 5, and 1.5 kbp DNA. Threading time dependence on DNA length for events recorded at 250 mV (right panel).