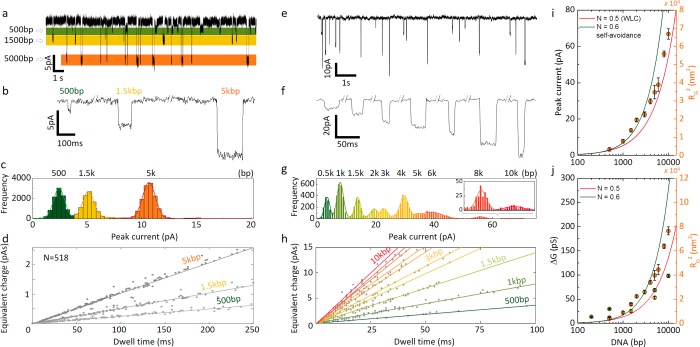
Figure 5.
Detection of mixed dsDNA sample in the nanobridge configuration. (a) Translocation signals of a sample containing 500 bp, 1.5 kbp, and 5 kbp at a concentration of 100 pM each in 100 mM KCl buffered in TE (pH 8.0) at 200 mV. (b) Representative current blockade traces of 500 bp, 1.5 kbp, and 5 kbp DNA. (c) Peak current histogram for a mixture containing 500 bp, 1.5 kbp, and 5 kbp. The mean peak current was obtained via Gaussian fitting (2.4 ± 0.5 pA for 500 bp, 5.1 ± 0.5 pA for 1.5 kbp, and 10.7 ± 0.6 pA for 5 kbp). (d) Equivalent charge plot was used to identify the different DNA population and was shown to be linear dependent on dwell time. The calculated slopes were 2.5 pA for 500 bp, 5.2 pA for 1.5 kbp, and 10.3 pA for 5 kbp. (e) Translocation signal of a 1 kbp DNA ladder, containing 10 DNA fragments (500 bp, 1.5 kbp, 2 kbp, 3 kbp, 5 kbp, 6 kbp, 8 kbp, and 10 kbp) at a total concentration of 100 pM in 100 mM KCl buffered in TE (pH 8.0) at 350 mV. (f) Representative current blockades and (g) peak current histogram. (h) Equivalent charge plot for the same sample as shown in g. (i) Peak current and (j) conductance for the 10 DNA fragments in the 1 kbp ladder (orange), sample from panel a (yellow), and data from Figure 4c (green). The scaling is in excellent agreement with the DNA radius of gyration squared (right axes) using a worm-like chain (WLC) model with and without self-avoidance.