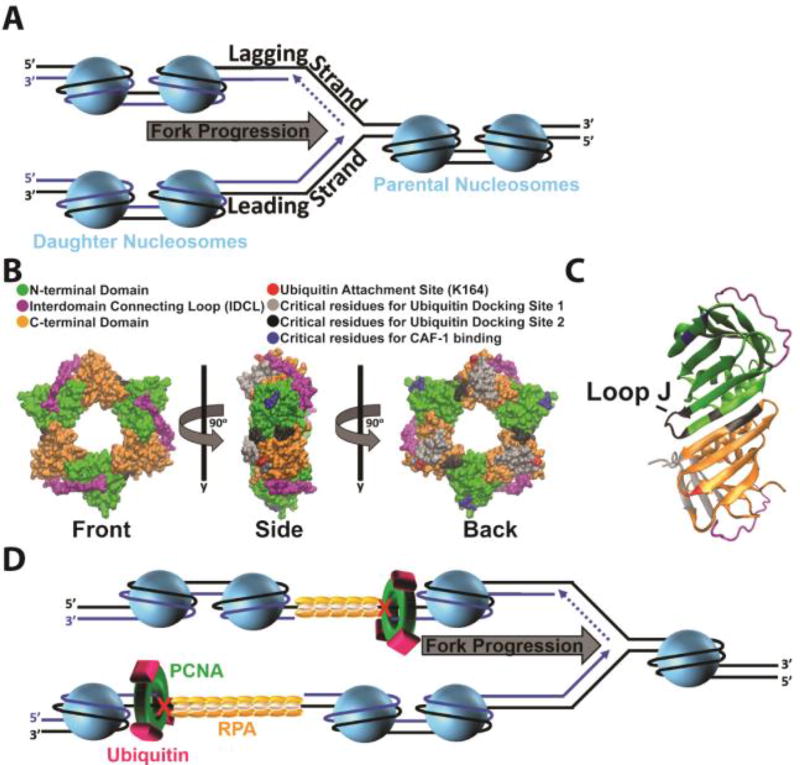
Figure 11.
Accessibility of postreplicative gaps. (A) Chromatin dynamics during S-phase. On the lagging strand template, Okazaki fragments yet to be ligated are indicated by dashed lines. During S-phase, nucleosomes ahead of a replication fork are disassembled as the replication fork progresses. Immediately following passage of a replication fork, nucleosomes are re-formed on the nascent daughter DNA by various histone chaperones. B) Monoubiquitination of PCNA and CAF-1 binding. Surface of the human PCNA ring generated with VMD (PDB 1AXC). Coloring scheme is indicated. Each PCNA monomer consists of two independent domains joined by an interdomain connecting loop (IDCL). Three PCNA monomers arrange in a head-to-tail manner, resulting in a ring with structurally distinct faces. The “front” face contains the IDCLs and interacts with pols. Following UV irradiation, single ubiquitin moieties are covalently attached to PCNA rings at the conserved lysine residue 164 (K164). C) Ubiquitin docking site 2. Shown in cartoon form is a side profile of a PCNA subunit-subunit interface. Coloring scheme is identical to panel B. At the interface, the N-terminal domain from one PCNA monomer interacts with the C-terminal domain from the adjacent monomer. At ubiquitin docking site 2, a ubiquitin moiety conjugated to K164 interacts with loop J (indicated) and residues within the subunit-subunit interface. (D) Selective inhibition of CAF-1 activity at postreplicative gaps. Upon formation of postreplicative gaps within either template, chromatin assembly continues in the wake of a progressing replication fork. PCNA is retained at a postreplicative gap and is monoubiquitinated by Rad6/Rad18. We propose that the ubiquitin moieties dock onto the PCNA ring at ubiquitin docking site 2, sheltering/disrupting the CAF-1 binding sites on PCNA. This precludes CAF-1 binding to PCNA, selectively inhibiting nucleosome assembly at/near a postreplicative gap until the gap is filled.