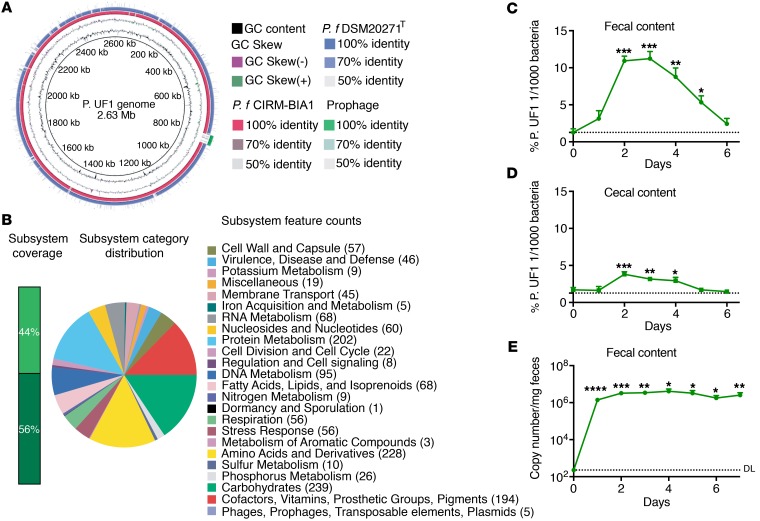
Figure 2. Characterization of P. UF1 bacterium.
(A) Genome sequence comparison of isolated P. UF1 with known P. freudenreichii spp. Shermanii CIRM-BIA1 (red) and P. freudenreichii spp. freudenreichii DSM20271T (blue). Prophage was identified by PHAST (green). Sequence identity between P. freudenreichii strains and P. UF1 is shown. (B) Correlation of the annotated genome sequence on the RAST platform with different metabolic pathways of P. UF1. Numbers in parentheses indicate numbers of annotated genes belonging to subcategories of pathways. (C and D) C57BL/6 mice (n = 3) were gavaged with P. UF1 (109 CFU/mouse) 1 time, and mice were sacrificed every day to detect P. UF1 in the fecal (C) and cecal (D) contents. (E) GF mice (n = 5) were gavaged with P. UF1 (109 CFU/mouse) 1 time, and fecal samples were collected every day to detect P. UF1. DL indicates the qPCR detection limit. Asterisks indicate statistical significance compared with day 0. Data are representative of 1 (E) or 3 (C and D) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 2-tailed unpaired t test (C–E).