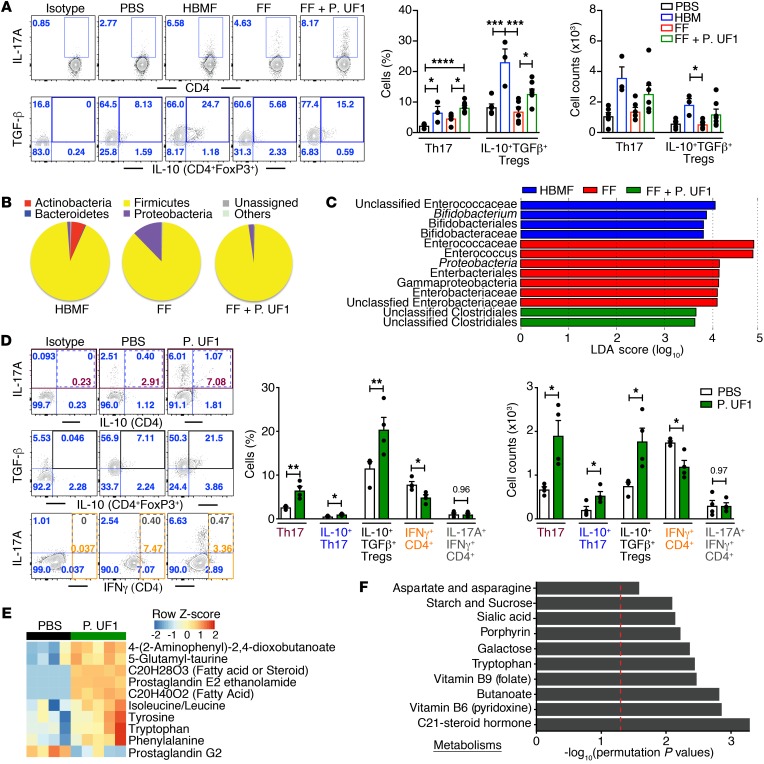
Figure 3. Modulation of colonic immune responses by HBMF preterm infants’ microbiota.
(A–C) GF mice were transfaunated with HBMF preterm infants’ microbiota (blue), FF preterm infants’ microbiota (red), or FF preterm infants’ microbiota plus 4 treatments with P. UF1 (green) or were left untransfaunated (black). Colonic immune responses and microbiota were analyzed 2 weeks later. Representative data of flow plots, percentages, and total cell counts of Th17 cells and IL-10+TGF-β+ Tregs (A). Microbial phyla structure (B) and LDA analysis (C) of fecal samples from indicated group. (D) GF mice were monoassociated with P. UF1 (green) or gavaged with PBS (white), and induced colonic immune responses were analyzed. Representative data of flow plots, percentages, and total cell counts of Th17 cells, IL-10+TGF-β+ Tregs, and IFN-γ+CD4+ cells (Th1 cells). (E and F) Heatmap of selected metabolites differentiating fecal samples from PBS- and P. UF1–gavaged GF mice (E), and significant metabolic pathways of fecal samples from PBS- versus P. UF1–gavaged GF mice (F). Red dashed line shows permutation of P = 0.05. Data are pooled from 2 independent experiments (n = 3–7 mice/group, A–C) or representative of 3 (n = 4–5 mice/group, E and F) or 6 (n = 4 mice/group, D) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ANOVA plus Tukey’s post test (A) or 2-tailed unpaired t test (D).