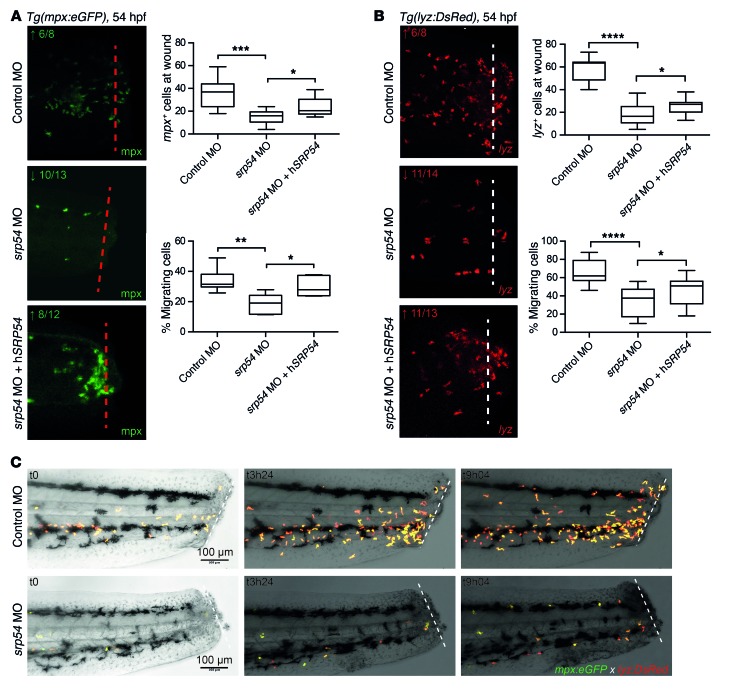
Figure 5. srp54 gene knockdown reduces neutrophil chemotaxis in zebrafish embryos.
(A and B) Tail fin wounding experiments and detection of neutrophils using anti-mpx immunostaining (A) and Tg(lyz:DsRed) embryos (B), respectively. Shown are representative images and corresponding quantitation of neutrophils, indicating reduced numbers at the wound in srp54 morphants compared with control-injected or srp54 MO plus hSRP54 mRNA–coinjected embryos. Dotted lines indicate the localization of the wound. Graphs summarize the counts of all visible neutrophils in the region of the wound and, respectively, the percentages of migrating neutrophils to the wound region (neutrophils at the wound relative to total numbers in the tail region, from the yolk sac extension to the tip of the wounded tail). Arrows indicate downregulation (↓) or rescued phenotype (↑) for each gene. Numbers indicate the sum of embryos with the respective phenotype/total number of embryos analyzed. Data from 3 biological replicates with 2 or more animals per group for each individual biological replicate experiment are shown. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05, by 1-way ANOVA with multiple comparisons. Error bars indicate the mean ± SD. (C) Still images of confocal time-lapse live imaging of tail fin amputation in Tg(mpx:eGFP lyz:DsRed) transgenic embryos. Shown are 3 representative time points for control-injected and spr54 MO–injected transgenic embryos, in which the migration of double-positive neutrophils to the wound area was visible over time. For each time point, merged images are shown. Dotted lines indicate the localization of the wound. Scale bars: 100 μm (A–C).