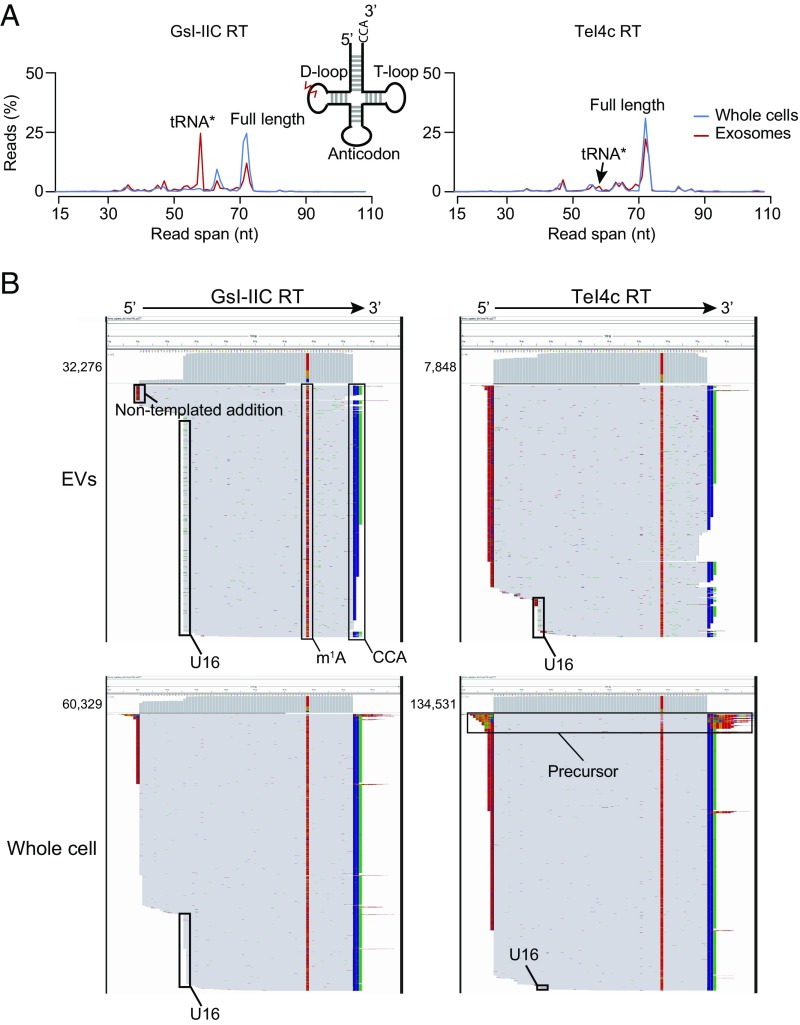
Fig. 7.
Evidence for an exosome-specific tRNA modification. (A) Read-span distributions for all reads mapping to tRNAs in TGIRT-seq libraries prepared from unfragmented whole-cell RNA and exosomal RNA with either GsI-IIC RT (TGIRT-III; Left) or TeI4c RT (Right). The peaks corresponding to full-length mature tRNA and the tRNA* species with a strong stop at position 16 in the D-loop are indicated. The Inset shows a tRNA schematic indicating the approximate position of the putative exosome-specific tRNA modification (jagged red line). (B) IGV screen shots showing read coverage across the tRNA coding sequence for a tRNA lysine isoacceptor (CTT) from TGIRT-seq of EV and unfragmented whole-cell RNAs with GsI-IIC RT or TeI4c RT. Coverage plots are above read alignments to the tRNA coding sequence with reads sorted by top strand position. The numbers of reads are indicated to the left of the coverage plot and were down sampled to 1,000 reads in IGV for visualization. Nucleotides in reads matching nucleotides in the annotated reference are colored gray. Nucleotides in reads that do not match the annotated reference are color coded by nucleotide (A, green; C, blue; G, brown; and T, red). 1-methyladenine (m1A) and the 3′ CCA posttranscriptional modifications, extra nontemplated nucleotides added to the 3′ end of the cDNA by the RT, and the D-loop modification (stop at position U16) are indicated. The reads identified as tRNA precursors have short 5′- and 3′-end extensions that map to genomic loci encoding this tRNA (SI Appendix, Fig. S14B).