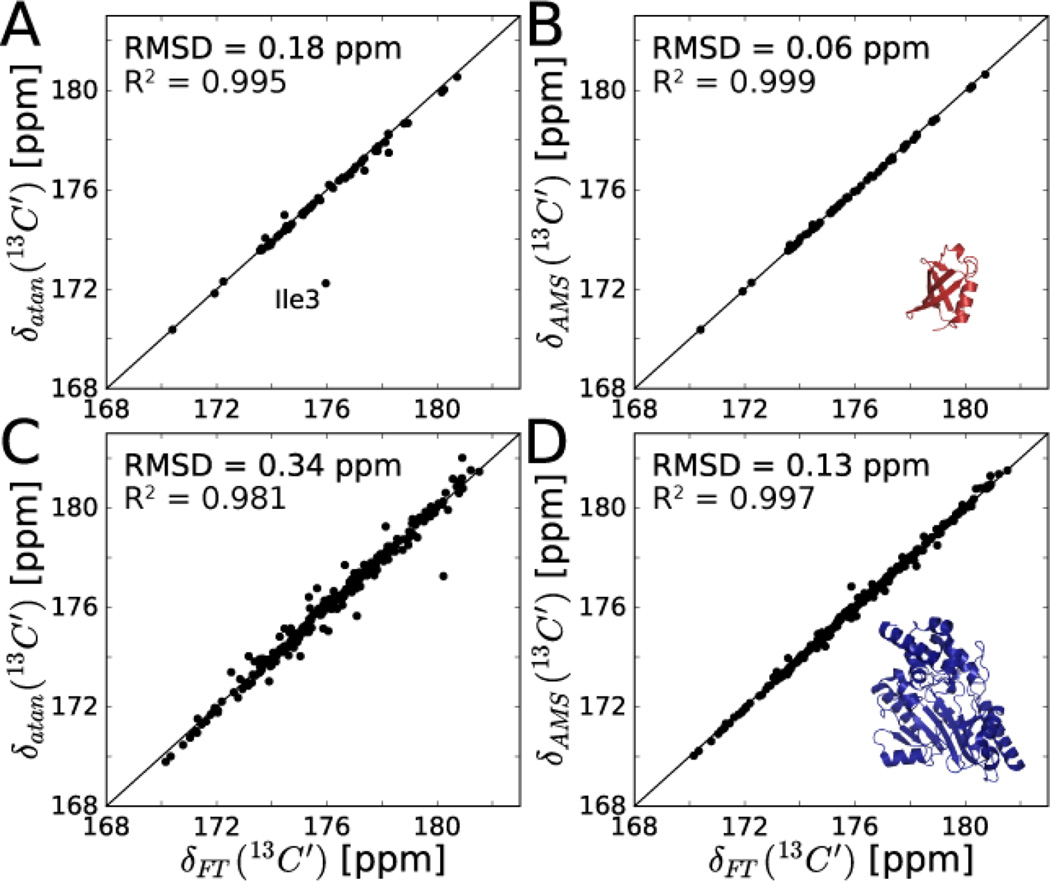
Figure 1.
Accuracy of arctangent approach (A,C) and AMS (B,D) for carbonyl chemical shift determination for 13C,15N labeled ubiquitin (A,B) and arginine kinase (C,D) from 3D HNCO data sets. In A, the outlier (Ile3) was excluded from RMSD and R2 calculations. Carbonyl shifts in A,C were determined from Eq. (1) using just the 2nd complex t1 time point of the HNCO. AMS-derived shifts in B,D are the best fit result to Eq. (2) using the first 4* t1 points of the HNCO. Reference chemical shifts δFT (x-axes) were determined by peak-picking the conventionally FT processed 3D HNCO experiments acquired with 40* and 44* t1 points for ubiquitin and arginine kinase, respectively.