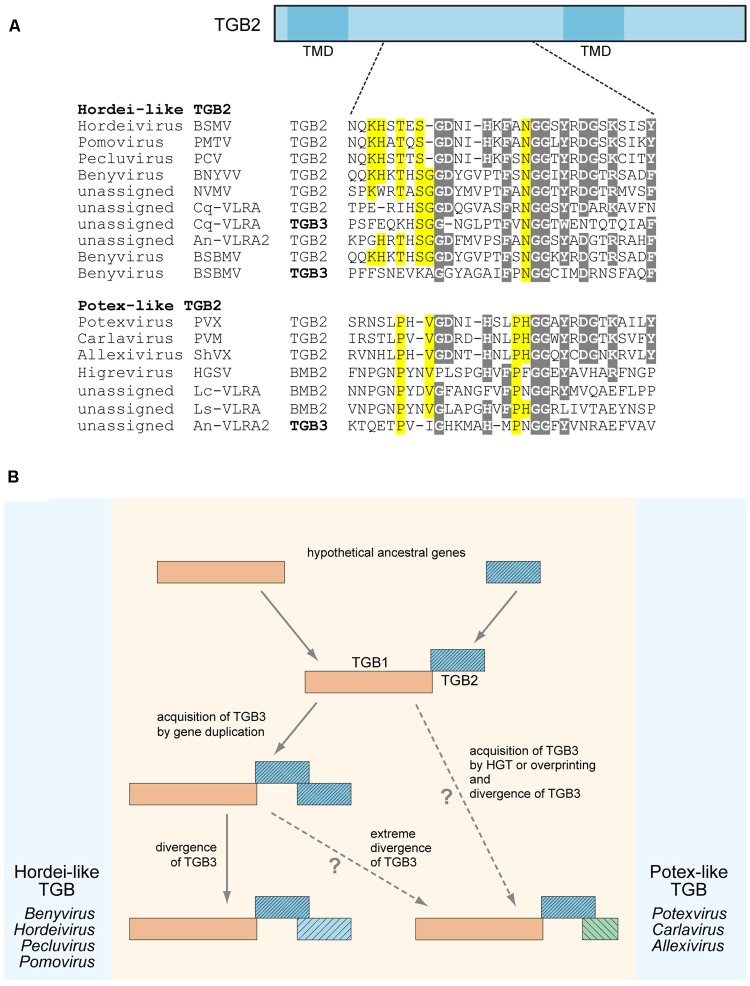
FIGURE 1.
Sequence conservation and possible evolutionary origin of TGB2 and TGB3 proteins. (A) Alignment of amino acid sequences located in the central conserved hydrophilic region between two transmembrane domains (TMD) of TGB2 proteins. TGB2 protein is schematically shown above the alignment as a box; the positions of TMDs are indicated; the position of aligned sequence region in TGB2 is shown by dashed lines. TGB3 sequences showing similarity to TGB2 sequences in the aligned region are included into the alignment and shown by bold. Alignments for hordei-like and potex-like TGB2 proteins are presented separately. Gray shading indicates residues conserved in both TGB types, yellow shading shows residues specifically conserved either in potex-like TGB2, or in hordei-like TGB2. BSMV, Barley stripe mosaic virus; PSLV, PMTV, Potato mop-top virus; BSBV, Beet soil-borne virus; PCV, Peanut clump virus; BNYVV, Beet necrotic yellow vein virus; NVMV, Nicotiana velutina mosaic virus; BSBMV, Beet soil-borne mosaic virus; PVX, Potato virus X; PVM, Potato virus M; ShVX, Shallot virus X; HGSV, Hibiscus green spot virus; CqVLRA, Colobanthus quitensis contig 125488 (NCBI accession GCIB01126289); An-VLRA2, Asplenium nidus assembly (1KP database accession PSKY-2058768); Lc-VLRA, Litchi chinensis assembly (1KP database accession WAXR-2010981); Ls-VLRA, Lathyrus sativus (NCBI accession GBSS01016353). (B) Proposed general scheme of TGB evolution. Processes specific for potex-like and hordei-like TGBs are shown on the right and left, respectively.