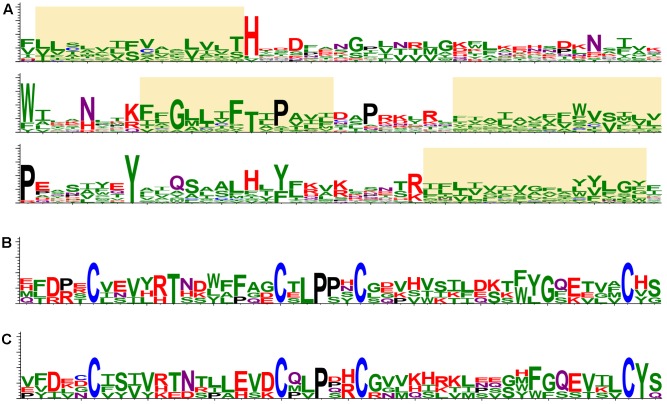
FIGURE 2.
Protein structure conservation depicted as sequence logos based on aligned protein sequences. (A) The most conserved region of SP24 proteins in plant and insect viruses. Yellow boxes indicate stretches of hydrophobic amino acid residues. The logo is based on the following sequences: Loreto virus (accession number KX518775); Anopheline-associated C virus (KF298279); Blueberry necrotic ring blotch virus (JN651150); Chronic bee paralysis virus (ANG65715); Hibiscus green spot virus (HQ852054); Ambrosia trifida (GEOH01007094); Triticum polonicum (GEDQ01066052); Camellia sinensis (GFMV01045386 and GARM01000026); Paulownia tomentosa (GEFV01018861); Panax ginseng (GDQW01045297); Elaeocarpus photiniifolius (FX134396); Gevuina avellana (GEAC01035000); Citrus leprosis virus C (NC008170); Humulus lupulus (GAAW01021049). (B) CBPV ORF2-like proteins in insect viruses. The logo is based on the following sequences: Chronic bee paralysis virus, Loreto virus, Negev virus, Piura virus, and Wuhan house centipede virus 1. (C) CBPV ORF2-like proteins in virus-like RNA in plant transcriptomes. The logo is based on the following sequences: Triticum polonicum (GEDQ01066052), Panax ginseng (GDQW01045299), Camellia sinensis (GFMV01053147), and Talbotia elegans (SILJ-2021063). The sequence logos visualize the distribution of amino acid residues at each position of conserved motifs. Amino acids are represented in a single-letter code and colored as follows: N and Q, purple; D, E, K, R and H, red; P, black; C, blue; S, T, Y, A, I, L, M, F, V, G and W, green.