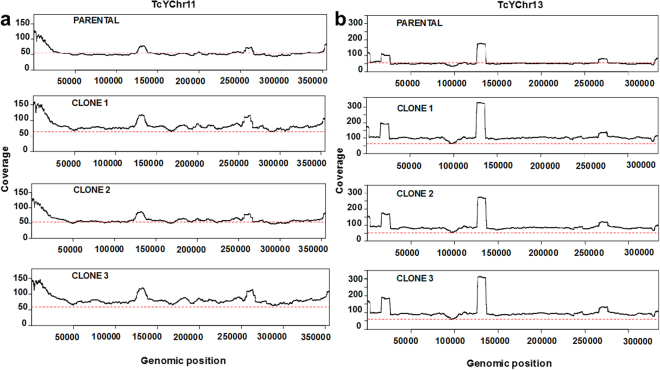
Figure 2.
Large-scale DNA copy-number variation in benznidazole-resistant clones. Histograms showing read-coverage for specific regions of chromosome 11 (a) and 13 (b) in parental and drug-resistant clones. Chromosome annotation was performed by ordering the contigs against the T. cruzi CL Brener reference genome21. High read peaks correspond to repetitive hypothetical protein genes, tandem repeats within kinesin-like genes (chromosome 11), linked histone H2A genes and short interspersed DNA sequences (chromosome 13). The red dotted line represents the average read-coverage across all chromosomes (single-copy genes). Assessment by the program control-freeC is consistent with an additional copy (trisomy) of the regions shown in chromosome 11 [from TCRUZ.110005800 (TcCLB.509349.30) to TCRUZ.110020000 (TcCLB.506443.80)] and chromosome 13 [from TCRUZ_130005000 (TcCLB.510165.60) to TCRUZ_130019000 (TcCLB.508325.90)]. Chromosome 11 in clone 2 did not display an increased read-coverage compared to the genome-wide average.