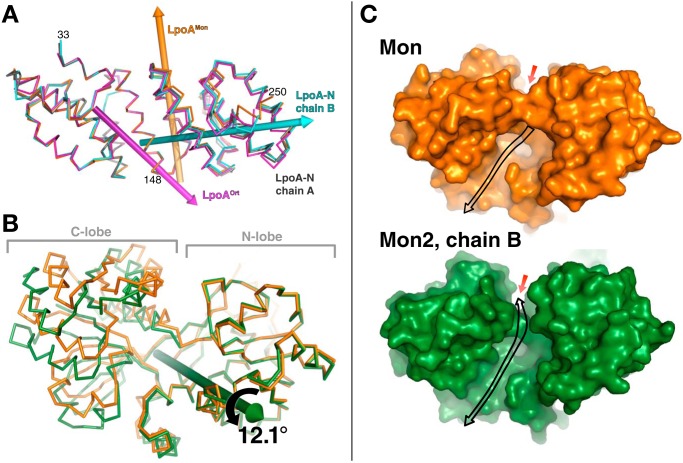
Figure 6.
Comparison of LpoA domain structures from different crystal forms. A, N domain twists and flexes. Overlay of the N domains of chain B of LpoA-N (cyan), LpoAOrt (magenta), and LpoAMon (orange) on that of chain A of LpoA-N (gray), based on least-squares fittings of their subdomain 1 (residues 33–149, left half of the figure). Rotations of the first three N-domain structures by about 4.7, 4.0, and 4.3°, respectively, around the correspondingly colored rotation axes yielded the best superpositions of their subdomains 2 (residues 150–248, right half of the figure) onto that of chain A of LpoA-N. This rotation axis in LpoAOrt (magenta) passes through residue 148 and is approximately parallel to H5, whereas that in LpoAMon (orange) is almost parallel to H8. The rotation axis for chain B of LpoA-N (cyan) runs through the center of subdomain 2 and is almost perpendicular to the LpoAMon axis. B, hinge bending in C domain. Shown is an overlay of the C domain of LpoAMon2 chain B (green) on that of LpoAMon (orange), after fitting their N lobes (residues 360–488 and 560–573) to each other. The C lobe of the LpoAMon2 chain B C domain was observed to be rotated, here by 12.1° around the green axis, with respect to the LpoAMon C lobe. Supplemental Fig. S2 shows two other C-domain comparisons. C, the consequence of the two lobes of the C domain spreading apart in B is that the LpoAMon2 chain B (bottom; green solvent contact surface) has a more direct pathway (black arrow) to the electronegative region on top of the C domain (red arrow; also see Fig. 4E). In contrast, in the LpoAMon (top, orange) C-domain structure, access to the electronegative region is blocked. This may affect the site accessibility to a bound polypeptide or PG strand.