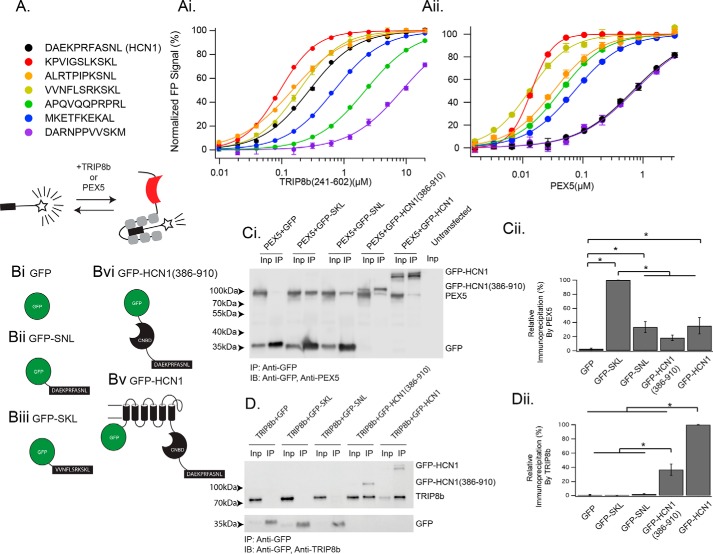
Figure 1.
The TRIP8b TPR domains bind a variety of peptide ligands. A, legend showing the sequence of the peptides used as well as a schematic for the experimental paradigm. Ai, TRIP8b241–602 was titrated into 50 nm of the indicated TAMRA-labeled peptide, with binding detected as a change in fluorescence polarization. See supplemental Table 1 for Kd values. Aii, the same experiment as in Ai was performed with PEX5 substituted for TRIP8b241–602. Error bars represent S.D. for a single run of the experiment, which was performed on three separate occasions and in triplicate on each occasion. Bi–Biv, schematics showing the GFP-tagged constructs used in C and D. Ci and Cii, co-IP experiments were performed with HEK293 cells transfected with the indicated constructs (see “Experimental procedures” for details of the volume loaded in each lane). In each case, GFP was immunoprecipitated, and then PEX5 and GFP were immunoblotted (IB). Note that GFP-HCN1(386–910) is very close in size to PEX5 and appears as the band above the PEX5 band. Also of note is the anomalous migration of the peptide-labeled GFP constructs. Based on our sequencing results, we are confident that this discrepancy is the result of a change in the migration of the protein in the SDS-PAGE gel secondary to the addition of a peptide tag to the C terminus (52). As described in the text, only GFP-SKL is efficiently bound by PEX5. Quantification is provided in Cii and normalized by the interaction between PEX5 and GFP-SKL (one-way ANOVA, F(4,19) = 32.91; GFP-SKL, 100% ± 0%; GFP-SNL, 33.3% ± 7.8%; GFP-HCN1(386–910), 18.4% ± 3.8%; GFP, 2.5% ± 1.1%; GFP-HCN1, 35.4% ± 11.49%; n = 4 distinct experiments). Inp, input. Di, an identical experiment as in C was performed, with the exception that TRIP8b was substituted for PEX5. Note that GFP-SKL fails to bind TRIP8b, whereas both GFP-HCN1(386–910) and GFP-HCN1 efficiently bind TRIP8b. Dii, quantification of the results provided in Di. TRIP8b elutions were scaled to the input fraction and normalized by the interaction between TRIP8b and GFP-HCN1 (see “Experimental procedures”) (F(4,19 = 146.69; GFP-HCN1, 100% ± 0.0%; GFP-HCN1(386–910), 36.6% ± 7.9%; GFP-SNL, 2.1% ± 0.0%; GFP-SKL, 0.2% ± 0.3%; GFP, 0.8% ± 0.6%; n = 4 distinct experiments). See supplemental Fig. 2 for an alternative display setting of the blot in D that highlights the band in the GFP-SNL IP lane, which is nonzero but statistically insignificant. *, p < 0.05 by Tukey's HSD post hoc test.