Abstract
It is increasingly appreciated that missense variants may not only alter protein structure and function but may also influence pre-mRNA splicing and/or mRNA stability. Here we explore this issue in the context of currently known SPINK1 missense variants using a full-length gene assay. We demonstrated that 4 (17%) out of 24 variants tested significantly reduced pre-mRNA splicing and/or stability as compared with the wild-type. However, since the strongest effect observed was a 23% reduction from normal, the contribution of SPINK1 missense variants to the clinical phenotype through an impact on mRNA processing alone may be relatively minor compared with their effects in relation to protein structure/function.
Keywords: chronic pancreatitis, full-length gene assay, minigene assay, missense variant, mRNA stability, pre-mRNA splicing, SPINK1 gene
1. Introduction
Recently, it was reported that that up to 10% of known disease-associated missense variants alter pre-mRNA splicing [1]. In addition, missense variant-containing transcripts may also be less stable as compared to wild-type, if for example the variants facilitate the formation of alternative mRNA secondary structures [2]. SPINK1 (encoding pancreatic secretory trypsin inhibitor; OMIM #167790) is one of the most studied genes predisposing to chronic pancreatitis [3]. We have recently provided both in vitro and in silico evidence [4] against a significant effect of the SPINK1 c.194G>A variant on pre-mRNA splicing, contrary to previous claims by Beer and Sahin-Tóth [5]. The key difference between these studies was that the former used a full-length gene assay to assess pre-mRNA splicing, whereas the latter used a minigene assay; full-length gene assays are generally superior to minigene assays because pre-mRNA splicing is highly dependent upon the genomic context [6]. We were thus able to conclude that the pathogenic effect of the c.194G>A missense variant (p.Arg65Gln) was exerted exclusively via its impact on protein secretion, as previously reported [7,8], rather than through an influence on pre-mRNA splicing.
Prompted, however, by the aforementioned publication of Soemedi et al. [1], we sought to analyze the effect of all currently known SPINK1 missense variants on pre-mRNA splicing and/or mRNA stability by means of our previously established full-length gene assay [4].
2. Materials and Methods
2.1. Mutagenesis
The 23 additional SPINK1 missense variants as well as three synonymous variants logged in the Genetic Risk Factors in Chronic Pancreatitis database [9]; Table 1) were introduced separately into the pcDNA3.1/V5-His-TOPO vector harboring the wild-type full-length 7-kb SPINK1 genomic sequence (including all four exons plus the three introns of the gene [10]) with Agilent’s QuikChange II XL Site-Directed Mutagenesis Kit (Les Ulis, France), essentially as previously described [11]. The sequences of the mutagenesis primers are available upon request. The successful introduction of the desired variants was confirmed by direct sequencing; the primers used for this purpose included 5′-TAATACGACTCACTATAGGG-3′ (for exon 1 variants), 5′-TGCCTGATTCATTTCCAGCAG-3′ (for exon 2 variants), 5′-TCAGAAGGGCCATAGGACTT-3′ (for exon 3 variants), and 5′-AGAGGCATCAGGAGCAAAAG-3′ (for exon 4 variants).
Table 1.
Quantitative reverse-transcription PCR (RT-PCR) analyses of mRNA expression from HEK293T cells transfected with SPINK1 missense variants.
| Exon | Variant | Relative mRNA Expression a | Pathogenicity Classification b | ||
|---|---|---|---|---|---|
| Nucleotide Change | Amino Acid Change | Mean ± SD | p-Value | ||
| Missense | |||||
| 1 | c.26T>G | p.Leu9Arg | 94.3 ± 8.0 | 0.248 | Unknown |
| 1 | c.29G>A | p.Ser10Asn | 80.0 ± 4.4 | 0.003 | Non-pathogenic |
| 1 | c.36G>C | p.Leu12Phe | 96.4 ± 9.8 | 0.517 | Non-pathogenic |
| 1 | c.41T>C | p.Leu14Pro | 91.5 ± 5.9 | 0.064 | Pathogenic |
| 1 | c.41T>G | p.Leu14Arg | 103.6 ± 12.9 | 0.621 | Pathogenic |
| 3 | c.101A>G | p.Asn34Ser | 103.8 ± 7.6 | 0.482 | Non-pathogenic |
| 3 | c.110A>G | p.Asn37Ser | 99.0 ± 6.2 | 0.800 | Non-pathogenic |
| 3 | c.123G>C | p.Lys41Asn | 97.0 ± 4.0 | 0.315 | Pathogenic |
| 3 | c.126A>G | p.Ile42Met | 108.2 ± 9.4 | 0.270 | Unknown |
| 3 | c.133C>T | p.Pro45Ser | 94.7 ± 6.0 | 0.266 | Unknown |
| 3 | c.137T>A | p.Val46Asp | 100.1 ± 2.7 | 0.954 | Unknown |
| 3 | c.143G>A | p.Gly48Glu | 106.0 ± 11.6 | 0.379 | Pathogenic |
| 3 | c.150T>G | p.Asp50Glu | 98.8 ± 5.0 | 0.700 | Pathogenic |
| 3 | c.160T>C | p.Tyr54His | 100.5 ± 15.7 | 0.953 | Pathogenic |
| 3 | c.163C>T | p.Pro55Ser | 105.9 ± 8.4 | 0.355 | Non-pathogenic |
| 3 | c.190A>G | p.Asn64Asp | 82.2 ± 4.1 | 0.017 | Pathogenic |
| 3 | c.193C>T | p.Arg65Trp | 97.8 ± 4.7 | 0.505 | Unknown |
| 3 | c.194G>A c | p.Arg65Gln | 108.5 ± 10.4 | 0.295 | Pathogenic |
| 4 | c.198A>C | p.Lys66Asn | 97.5 ± 4.4 | 0.425 | Pathogenic |
| 4 | c.199C>T | p.Arg67Cys | 77.1 ± 10.1 | 0.007 | Pathogenic |
| 4 | c.200G>A | p.Arg67His | 94.3 ± 9.7 | 0.323 | Pathogenic |
| 4 | c.203A>G | p.Gln68Arg | 87.4 ± 7.0 | 0.037 | Protective |
| 4 | c.206C>T | p.Thr69Ile | 106.8 ± 7.1 | 0.240 | Pathogenic |
| 4 | c.236G>T | p.Cys79Phe | 110.0 ± 4.5 | 0.062 | Pathogenic |
| Synonymous | |||||
| 2 | c.75C>T | p.Ser25= | 90.2 ± 8.1 | 0.093 | Unknown |
| 3 | c.174C>T | p.Cys58= | 96.6 ± 4.2 | 0.291 | Unknown |
| 4 | c.231G>A | p.Gly77= | 93.3 ± 8.3 | 0.297 | Unknown |
a Relative to the wild-type value, which was set to 100 (results from three to five independent transfection experiments). SD, standard deviation. Variants whose expression levels were statistically different from wild-type (i.e., p < 0.05) are highlighted in blue. b In accordance with the Genetic Risk Factors in Chronic Pancreatitis database [9]. c Previously analyzed by means of the full-length gene assay [4].
2.2. Cell Culture, Transfection, RNA Extraction, and Reverse Transcription
Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal calf serum. Twenty-four hours prior to transfection, 3.5 × 105 cells were seeded per well of 6-well plates. For qualitative reverse transcription-PCR (RT-PCR) analyses (see Section 2.3), 1 µg wild-type or variant SPINK1 expression vector, mixed with 3 mL Lipofectamine 2000 Reagent (Life Technologies, Courtaboeuf, France), was used for transfection per well. For quantitative RT-PCR analyses (see Section 2.4), 500 ng wild-type or variant SPINK1 expression vector plus 500 ng pGL3-GP2 minigene [10] were mixed with 3 mL Lipofectamine 2000 Reagent for transfection. Forty-eight hours after transfection, total RNA was extracted using the RNeasy Mini Kit (Qiagen, Courtaboeuf, France). Reverse transcription was performed with 1 μg total RNA and 20mer-oligo(dT) by means of the SuperScript III Reverse Transcriptase kit (Life Technologies) in accordance with the manufacturer’s instructions. The resulting complementary DNAs (cDNAs) were treated with 2U Ribonuclease H (Life Technologies) at 37 °C for 20 min to degrade the remaining RNA.
2.3. Qualitative RT-PCR Analyses and Sequencing of the Resulting Products
Qualitative RT-PCR was performed in a 25-μL reaction mixture containing 12.5 μL HotStarTaq Master Mix (Qiagen), 0.4 μM each primer (forward: 5′-GGAGACCCAAGCTGGCTAGT-3′, located within the 5′-untranslated region encoded by the pcDNA3.1/V5-His-TOPO vector; reverse: 5′-AGACCGAGGAGAGGGTTAGG-3′, located within the V5 epitope sequence encoded by the pcDNA3.1/V5-His-TOPO vector) and 1 μL cDNA. The PCR program had an initial denaturation at 95 °C for 15 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 58 °C for 45 s, extension at 72 °C for 5 min, with a final extension step at 72 °C for 10 min. PCR products were cleaned by ExoSAP-IT (Life Technologies) and sequenced using the BigDye Terminator v1.1 Cycle Sequencing Kit (Life Technologies), the primer used for sequencing was the aforementioned 5′-GGAGACCCAAGCTGGCTAGT-3’.
2.4. Quantitative RT-PCR Analyses
These were performed essentially as previously described [10,12]. In brief, wild-type or variant SPINK1 expression vector was co-transfected with the previously constructed pGL3-GP2 minigene [10]. The target (SPINK1) and reference (i.e., pGL3-GP2 minigene) genes were PCR-amplified separately; SPINK1 expression was determined using pGL3-GP2 minigene expression as a reference by means of the ΔΔCt method in accordance with Pfaffl’s efficiency-corrected calculation model [13].
Quantitative RT-PCR was performed in a 25 µL mixture containing 12.5 µL HotStarTaq Master Mix Kit (Qiagen), 1 µL 1:25 diluted cDNA, 0.5 µM SYTO9 (Life Technologies), and 0.3 µM each primer; the primers for qualitative RT-PCR analyses of the SPINK1 gene (see Section 2.3) were used for analyzing the SPINK1 gene whilst the primers used for analyzing the reference gene were 5′-ACTGTTGGTAAAGCCACCAT-3′ (forward) and 5’-TGTATCTTATCATGTCTGCTCGAA-3’ (reverse). The PCR program had an initial denaturation step at 95 °C for 15 min, followed by 40 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. Quantitative analysis of each RT-PCR amplification was performed in triplicate on a Lightcycler 480II (Roche, Paris, France) using ‘relative quantification’ and ‘second derivative maxima’ options.
The difference between the expression levels of a SPINK1 variant and the wild-type sequence (results from three to five independent transfection experiments) was assessed for significance by the Student t-test. The difference was regarded as being statistically significant when the p-value was ≤0.05.
3. Results and Discussion
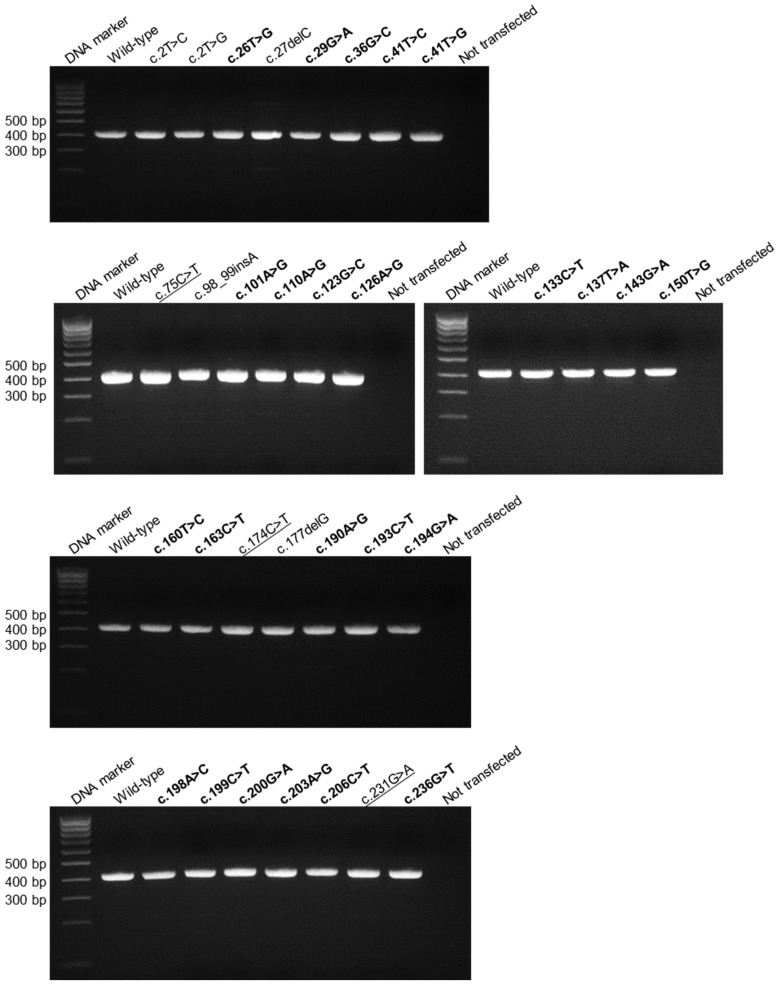
Qualitative RT-PCR analyses of mRNAs from HEK293T cells transfected with the 23 missense variants and 3 synonymous variants invariably yielded a single band of similar size to that of the wild-type (Figure 1). Sequencing of the 26 corresponding RT-PCR products revealed that all were correctly spliced as per the wild-type. When quantitative RT-PCR analyses were performed, a statistically significant lower mRNA expression level was observed with 4 of the 23 missense variants but not with any of the synonymous variants (Table 1). In this regard, it is pertinent to point out that the reliability of our quantitative RT-PCR method had been experimentally validated in our previous study [4].
Figure 1.
Qualitative RT-PCR analyses of HEK293T cells transfected with the full-length gene expression constructs carrying various SPINK1 exonic variants. The missense and synonymous variants under study (Table 1) are highlighted in bold and underlining, respectively. The remaining variants, all of which are clearly loss-of-function variants (i.e., translational initiation codon or frameshifting variants), were not discussed in this study. Note that the previously studied c.194G>A variant [4] was also included for the reason of completeness.
The decrease in mRNA expression noted for the four missense variants may be explicable in terms of two non-mutually exclusive mechanisms. Firstly, these variants could have led to the production of aberrantly spliced transcripts but these aberrant transcripts may have been subjected to rapid degradation by cellular defense mechanisms [14], rendering them undetectable by qualitative RT-PCR analysis. Secondly, and as mentioned above, correctly spliced variant-containing transcripts may be less stable as compared to wild-type through for example the formation of alternative mRNA secondary structures [2]. Whichever, the two different mechanisms are indistinguishable by our quantitative RT-PCR analysis.
Irrespective of the underlying biological mechanisms responsible, our systematic mRNA expression analysis of known SPINK1 missense variants is suggestive of the existence of a new layer of functional deficiency, at least in the case of two specific missense variants (i.e., c.190A>G (p.Asn64Asp) and c.199C>T (p.Arg67Cys)), the pathogenicity of which had already been established (Table 1). (N.B. The pathogenicity classifications of all 24 SPINK1 missense mutations (Table 1) were based essentially upon the analysis of in vitro-expressed mutant proteins in terms of their secretion and/or enzymatic activity.) Thus, the pathogenicity of these two variants may be seen to emanate from a combination of defective mRNA expression and protein function. By contrast, the other two missense variants characterized by a significant influence on pre-mRNA splicing and/or mRNA stability were previously classified as being either non-pathogenic or ‘protective’ (Table 1). Our new data may therefore prompt a re-consideration of these initial variant classifications. Specifically, the ‘non-pathogenic’ c.29G>A (p.Ser10Asn) variant may actually serve to increase the risk of chronic pancreatitis through its impact on mRNA expression and/or stability. As for the c.203A>G (p.Gln68Arg) variant, any ‘protective’ effect due to the significantly increased secretion of the mutant protein may be partly compensated for by its negative effect on pre-mRNA splicing and/or stability. This notwithstanding, two caveats should be borne in mind. First, the strongest observed effect on pre-mRNA splicing and/or stability was a 23% reduction from normal (c.199C>T). Whether such decreases are meaningful in a pathological context, particularly when we consider that the alleles responsible occur only in the heterozygous state in their respective carriers, is unclear. Second, the analyses should ideally be performed in a human pancreatic acinar cell line, which unfortunately is not yet available.
4. Conclusions
In summary, we demonstrate that four (~17%) of the currently known 24 SPINK1 missense variants exert a significant impact on pre-mRNA splicing and/or mRNA stability. However, since the strongest effect observed was a 23% reduction from normal, the contribution of SPINK1 missense variants to the clinical phenotype through their impact on mRNA processing is likely to be relatively minor. Finally, to the best of our knowledge, this study is the first to comprehensively analyze the consequences of all known missense mutations in a given gene on mRNA expression/stability in the context of a full-length gene assay. It remains to be seen if comparable results will be obtained for missense variants associated with other diseases.
Acknowledgments
H.W., who received a one-year scholarship from the China Scholarship Council (No. 201503170355), was a joint PhD student between the Changhai Hospital and INSERM U1078. Support for this study came from the National Natural Science Foundation of China (Grant Nos. 81422010 and 81470884; to Z.L.), the Shuguang Program of Shanghai (Grant No. 15SG33; to Z.L.), the Chang Jiang Scholars Program of Ministry of Education, China (Grant No. Q2015190; to Z.L.) and the Precision Medicine Program of the Second Military Medical University (Grant No. 2017JZ02; to Z.-S.L.); the Conseil Régional de Bretagne, the Association des Pancréatites Chroniques Héréditaires, the Association de Transfusion Sanguine et de Biogénétique Gaetan Saleun, and the Institut National de la Santé et de la Recherche Médicale (INSERM), France.
Author Contributions
J.-M.C. conceived and designed the experiments with the assistance of Z.-S.L., Z.L. and C.F. H.W. and A.B. performed the experiments. J.-M.C. wrote the paper. D.N.C. critically revised the manuscript. All authors analyzed the data and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Soemedi R., Cygan K.J., Rhine C.L., Wang J., Bulacan C., Yang J., Bayrak-Toydemir P., McDonald J., Fairbrother W.G. Pathogenic variants that alter protein code often disrupt splicing. Nat. Genet. 2017;49:848–855. doi: 10.1038/ng.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning K.S., Cooper T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017;18:102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegyi E., Sahin-Tóth M. Genetic risk in chronic pancreatitis: The trypsin-dependent pathway. Dig. Dis. Sci. 2017;62:1692–1701. doi: 10.1007/s10620-017-4601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H., Boulling A., Cooper D.N., Li Z.S., Liao Z., Chen J.M., Férec C. In vitro and in silico evidence against a significant effect of the SPINK1 c.194G>A variant on pre-mRNA splicing. Gut. 2017 doi: 10.1136/gutjnl-2017-313948. [DOI] [PubMed] [Google Scholar]
- 5.Beer S., Sahin-Tóth M. Exonic variants affecting pre-mRNA splicing add to genetic burden in chronic pancreatitis. Gut. 2014;63:860–861. doi: 10.1136/gutjnl-2013-305981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X.D., Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulling A., Le Maréchal C., Trouvé P., Raguénès O., Chen J.M., Férec C. Functional analysis of pancreatitis-associated missense mutations in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur. J. Hum. Genet. 2007;15:936–942. doi: 10.1038/sj.ejhg.5201873. [DOI] [PubMed] [Google Scholar]
- 8.Király O., Wartmann T., Sahin-Tóth M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut. 2007;56:1433–1438. doi: 10.1136/gut.2006.115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Genetic Risk Factors in Chronic Pancreatitis Database. [(accessed on 30 December 2016)]; Available online: http://www.pancreasgenetics.org/index.php.
- 10.Boulling A., Chen J.M., Callebaut I., Férec C. Is the SPINK1 p.Asn34ser missense mutation per se the true culprit within its associated haplotype? WebmedCentral Genet. 2012;3:WMC003084. [Google Scholar]
- 11.Zou W.B., Boulling A., Masson E., Cooper D.N., Liao Z., Li Z.S., Férec C., Chen J.M. Clarifying the clinical relevance of SPINK1 intronic variants in chronic pancreatitis. Gut. 2016;65:884–886. doi: 10.1136/gutjnl-2015-311168. [DOI] [PubMed] [Google Scholar]
- 12.Zou W.B., Boulling A., Masamune A., Issarapu P., Masson E., Wu H., Sun X.T., Hu L.H., Zhou D.Z., He L., et al. No association between CEL-HYB hybrid allele and chronic pancreatitis in Asian populations. Gastroenterology. 2016;150:1558–1560. doi: 10.1053/j.gastro.2016.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siwaszek A., Ukleja M., Dziembowski A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014;11:1122–1136. doi: 10.4161/rna.34406. [DOI] [PMC free article] [PubMed] [Google Scholar]