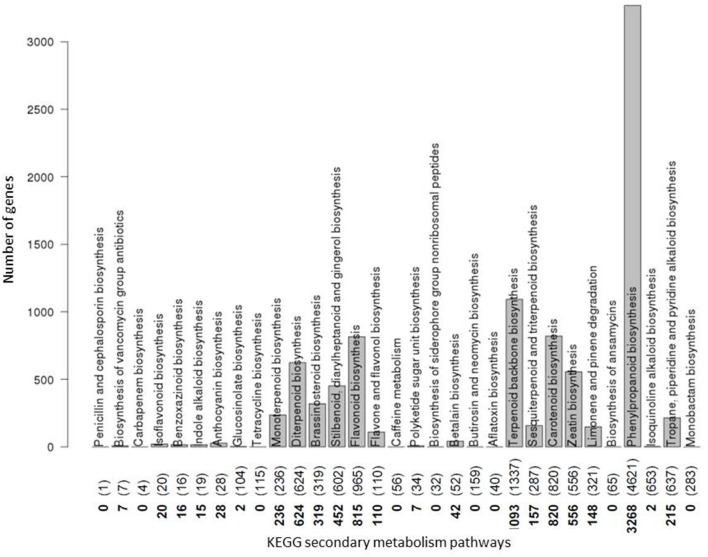
Figure 4.
Number of genes coding for enzymes involved in secondary metabolite pathways. The number of genes assigned to each plant secondary metabolite pathway as annotated in the KEGG database is plotted. Counts are based on all plant genes from any of the 24 plant species considered in this study. Numbers along the abscissa denote the actual number of genes involved in secondary metabolism pathways only with numbers in parentheses referring to the respective counts when considering genes annotated to participate in both secondary and primary metabolism pathways. Pathways are indicated by their names. Pathways “Biosynthesis of vancomycin group antibiotics” as a bacterial pathway, and “Penicillin and cephalosporin biosynthesis” as a fungal pathway were not considered any further in this plant-focused study. For the pathways “Benzoxazinoid biosynthesis” and “Isoquinoline alkaloid biosynthesis,” while listed in KEGG, conflicting or no annotations were contained in Biomart and, therefore, were discarded from further analysis. All remaining 19 secondary metabolism pathways with non-zero KEGG gene counts were considered further.