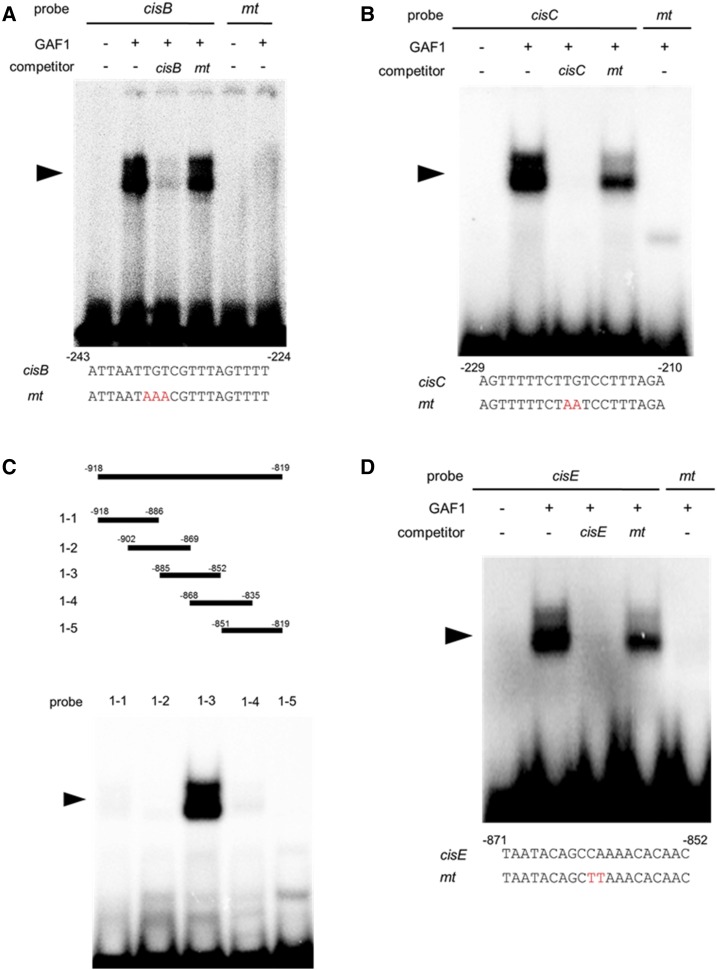
Figure 3.
Identification of GAF1-binding regions in the AtGA20ox2 promoter in vitro. A and B, Gel retardation assays using recombinant GAF1 protein. A, Oligonucleotides containing cisB (–243 to –224, wild type; lanes 1–4) or mtcisB (mt; lanes 5 and 6) were used as probes. B, Oligonucleotides containing cisC (–229 to –210, wild type; lanes 1–4) or mtcisC (mt; lane 5) were used as probes. Red letters indicate mutated bases. Wild type and mt indicate competition with a 200-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures. C, Gel retardation assay using recombinant GAF1 protein. Thirty-base-pair regions of the AtGA20ox2 promoter were used as probes. D, Oligonucleotides containing cisE (–871 to –852, wild type; lanes 1–4) or mtcisE (mt; lane 5) were used as probes. Red letters indicate mutated bases. Wild type and mt indicate competition with a 200-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures.