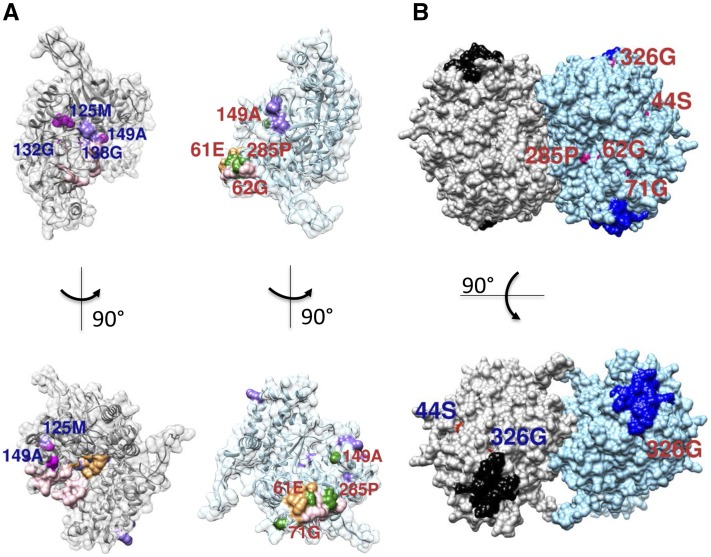
Figure 6.
Computation modeling and mapping of mutants on the GmSHMT08 structure. A, Mutation positions on or in close proximity to the functional sites (THF, MTHF, and FTHF in light pink, PLP-serine/PLP-glycine (PLS/PLG) in light orange, and Gly in light purple) for both interacting subunits. Shown in magenta are mutation positions for the first subunit and in green for the second subunit. B, GmSNAP18 protein-binding sites mapped on the GmSHMT08 tetramer together with its surface mutations. Shown are the surface mutations mapped on only one of the dimer subunits. The two GmSHMT08 dimers are colored gray and light blue. The two putative GmSNAP18 binding sites for each dimer are shown in black and navy. Shown in red (with blue labels) are mutation positions for one monomer of the first dimer and in magenta (with red labels) for a monomer of the second dimer.