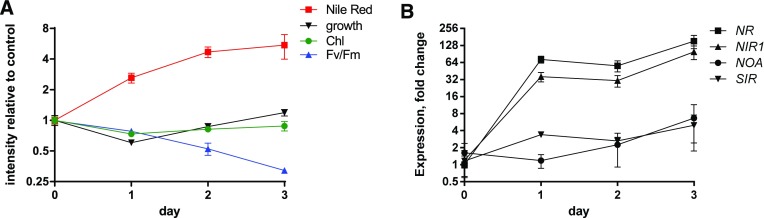
Figure 5.
Expression of NOA, NR, NIR, and SIR in nitrogen-depleted wild-type Phaeodactylum cells. Cells were harvested and resuspended in nitrogen-free media in 100-mL cultures a concentration of 106 cells · mL−1. Each day, a 20-mL culture aliquot was harvested for RNA extraction and a 300-µL culture aliquot was used for physiological measurements. A, Physiological parameters. Chl was measured by the absorption at 680 nm at room temperature, Nile red fluorescence was quantified to indicate neutral lipid contents, Fv/Fm was measured using fast chlorophyll fluorescence kinetics, and cell concentrations were estimated via the absorption at 730 nm. B, NOA, NR, NIR, and SIR gene expression. RNA was extracted from a cell pellet and reversely transcribed. Quantitative real-time PCR was conducted on 20 ng cDNA using oligonucleotides binding TUB as internal control and NOA, NR, and NIR as genes of interest. The SIR gene was used as a nitrogen-unrelated control. Data are normalized with value measured with cells harvested before the shift. Data are the results of three independent biological replicates.