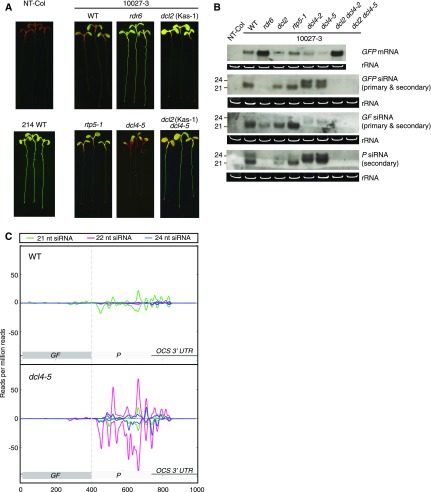
Figure 2.
dcl2 and dcl4 mutants display contrasting systemic PTGS phenotypes and levels of RDR6-dependent secondary siRNAs. Seedlings were grown in vitro under long days for 1 week. A, Typical GFP phenotypes of 10027-3 wild-type (WT) and mutant seedlings. All 10027-3 lines were homozygous for the 10027-3 T-DNA locus. Nontransgenic wild-type (NT-Col) and WT 214 (the wild-type genotype expressing a p35S:GFP transgene) seedlings are shown as GFP-negative and -positive controls, respectively. rdr6 and dcl2 (Kas-1) mutant and dcl2 (Kas-1) dcl4-5 double mutant seedlings are defective in systemic PTGS; by contrast, dcl4-5 seedlings showed enhanced systemic PTGS compared with wild-type seedlings. B, Northern blot of root GFP mRNA and GFP-, GF-, or P-specific siRNAs in nontransgenic wild-type (NT-Col) and 10027-3 genotypes. Both dcl4-5 and dcl4-2 mutant roots showed enhanced biogenesis of P-specific siRNAs, and both dcl2 dcl4-5 and dcl2 dcl4-2 double mutant roots were devoid of GFP-specific siRNAs. The GFP siRNA probe was the full-length 720-nucleotide GFP coding sequence. The GF and P siRNA probes were nucleotides 9 to 400 and 401 to 720 of the coding sequence, respectively. In A and B, dcl2 refers to the dcl2 (Kas-1) mutant and rdr6 refers to the sde1-1 allele. C, Enhanced accumulation of GFP-specific siRNAs in 10027-3 dcl4-5 shoots. The positive and negative scales on the y axis represents sense and antisense siRNA alignments, respectively. The x axis shows nucleotide positions in the GFP coding sequence and 3′ UTR. The data presented are averages of three independent biological replicates. The individual replicates are shown in Supplemental Figure S10.