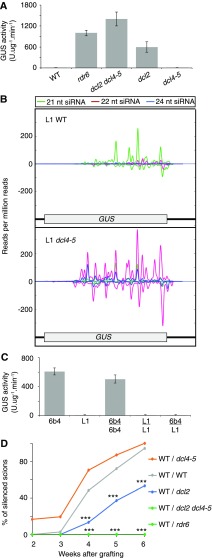
Figure 5.
DCL2 is required for RDR6-dependent, systemic PTGS of GUS. A, GUS activity in shoots of L1 genotypes, showing defective PTGS of p35:GUS in rdr6 and dcl2 mutants but not in dcl4-5. B, Contrasting GUS-specific siRNA profiles in 1-week-old roots of L1 wild-type (WT) and L1 dcl4-5 plants. Positive and negative scales on the y axis represent sense and antisense siRNA profiles, respectively. The x axis shows nucleotide positions in the GUS coding sequence and 3′ UTR. C, GUS activity in shoots of grafted and ungrafted 6b4 and L1 wild-type plants. 6b4 and L1 both carry a single p35S:GUS transgene but at different locations in the genome (Elmayan et al., 1998). For A and C, ungrafted and grafted plants were grown for up to 6 weeks in soil under long days. Ten to 20 plants were analyzed in each of two independent experiments (n = 2). Error bars represent se. D, Transmission of RDR6-dependent PTGS of GUS is compromised from dcl2 rootstocks but enhanced from dcl4 rootstocks. 6b4 wild-type scions were grafted onto L1 wild-type or mutant rootstocks. A piece of leaf was harvested each week after grafting for the measurement of GUS activity. The results are expressed as percentages of silenced scions. P values for pairwise comparison of each treatment with wild type/wild type are indicated (Fisher’s exact tests followed by Benjamini and Hochberg multiple correction: *, P < 0.05; **, P < 0.01; and ***, P < 0.001). For individual week 2 to 4 data, P values for dcl4-5 rootstocks compared with wild-type rootstocks were less than 0.1 but not significant (i.e. P > 0.05). However, pairwise comparison between wild-type and each mutant rootstock for combined week 2 to week 6 data generated highly significant P values of 3.67 × 10−10 (dcl2), 0.003 (dcl4-5), and 4.67 × 10−10 (rdr6 and dcl2 dcl4-5). In total, 100 to 243 grafted plants were assessed for each combination of genotypes. In A, C, and D, GUS activity was determined by 4-methylumbelliferyl-β-d-glucuronide assays (fluorescence units per minute per microgram of total proteins quantified by the Bradford protocol). dcl2 refers to the dcl2 (Kas-1) allele, and rdr6 refers to the sgs2-1 allele. All L1 and 6b4 lines were homozygous for the respective GUS reporter T-DNA locus.