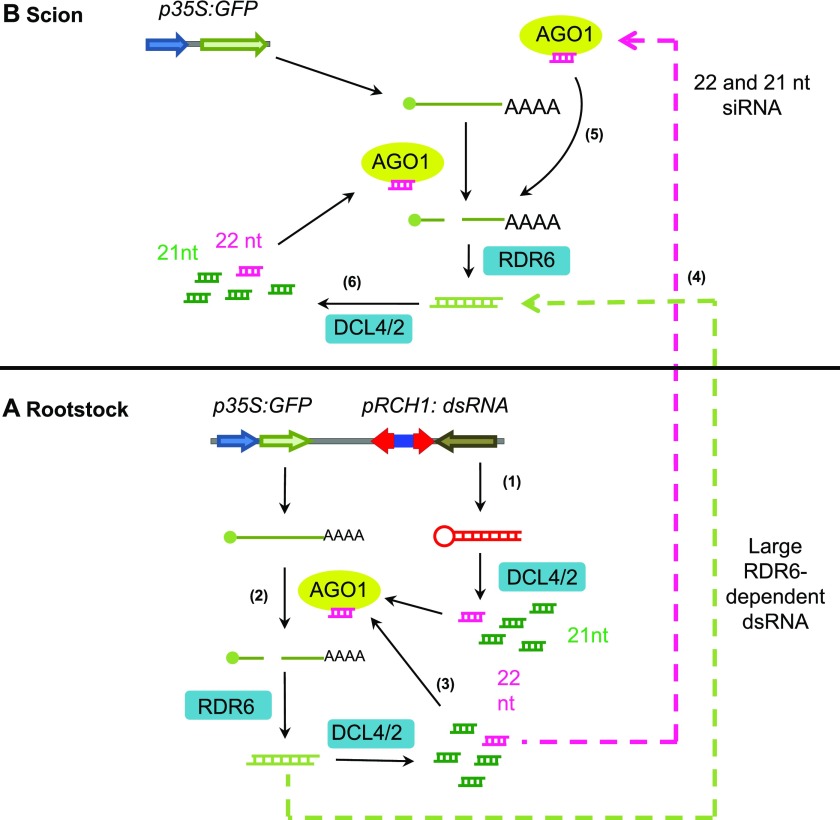
Figure 6.
Model for the production and reception of systemic PTGS signals in wild-type plants. A, GF-specific dsRNA produced in the root tip is processed into abundant 21-nucleotide (green) and low-abundance 22-nucleotide (pink) primary siRNAs by DCL4 and DCL2, respectively (step 1). These siRNAs are loaded into AGO1 and guide the cleavage of GFP mRNA. DCL2 activity more efficiently recruits RDR6, which produces dsRNA from the cleaved mRNA (step 2). The RDR6-dependent dsRNA is processed by DCL4 and DCL2 into secondary siRNAs, forming an amplification feedback loop to direct the cleavage of GFP mRNAs and the production of additional secondary siRNAs (step 3). The extent of engagement of the RDR6-dependent amplification loop, and the extent of PTGS, depend on the activity of DCL2 and its capacity to recruit RDR6, perhaps directly or via the biogenesis of 22-nucleotide siRNAs. RDR6-dependent 21- to 22-nucleotide siRNAs and/or large RNA act as mobile PTGS signals and move from the rootstock to the scion to induce systemic PTGS (step 4). It is unknown whether (1) siRNA or large RNA systemic PTGS signals move as double-stranded or single-stranded molecules, (2) the movement of systemic PTGS signals is by diffusion or is facilitated by an active process, or (3) further amplification of systemic PTGS signals occur en route to the scion (Gursanscky and Carroll, 2012). B, In the scion, mobile 21- and 22-nucleotide siRNAs coming from the rootstock could be loaded into AGO1 to direct the cleavage of GFP mRNA and the induction of PTGS. In this scenario, cleavage of GFP mRNA by AGO1 in complex with 22-nucleotide siRNAs would recruit RDR6 and induce PTGS more efficiently than cleavage guided by 21-nucleotide siRNAs (step 5). Alternatively, large RDR6-dependent systemic RNA signals could engage the RDR6-dependent amplification loop directly to induce PTGS (step 6).