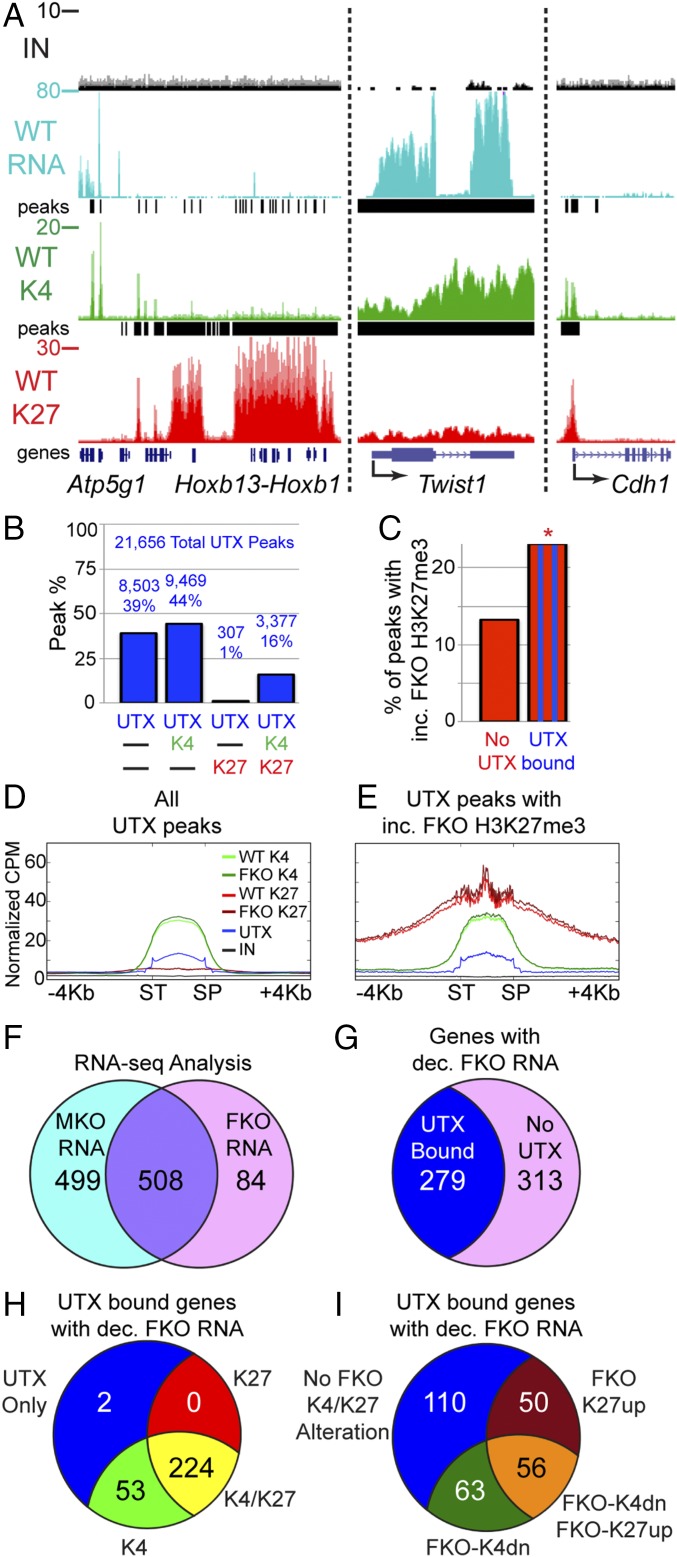
Fig. 4.
Demethylase-dependent and -independent UTX targets. (A) RNA-seq (RNA), H3K4me3 ChIP-seq (K4), and H3K27me3 ChIP-seq (K27) of isolated E12.5 WT cranial NC at indicated genes (IN, input; peaks, Chip-seq enrichment peaks). (B) Percentage of UTX peaks that overlap with H3K4me3 (K4) and/or H3K27me3 (K27). Total peak numbers are indicated. (C) Percentage of peaks with significantly increased (inc.) FKO H3K27me3 were significantly greater at sites overlapping with UTX binding compared with unbound H3K27me3 sites (23% vs. 13%, Fisher’s exact test *P < 0.001). (D and E) Profiles comparing WT to FKO across all scaled UTX peaks (D, SP, stop; ST, start) or UTX peaks that overlap with sites of increased (inc.) FKO H3K27me3 (E, same cpm scale). (F) RNA-seq identified 508 genes significantly reduced in both MKO and FKO expression relative to WT or 84 FKO specific genes (592 total). (G) The total set of 592 genes with decreased (dec.) FKO expression were overlaid with UTX-bound genes (UTX ChIP-seq peak within 10 kb of gene body) to identify direct UTX gene targets. (H) A total of 279 direct UTX targets were classified as having peaks of H3K27me3 (K27), H3K4me3 (K4), K4/K27, or UTX only within 10 kb of the gene body. (I) A total of 279 direct UTX targets were classified as experiencing enhanced FKO H3K27me3 (FKO-K27up), reduced FKO H3K4me3 (FKO-K4dn), both alterations, or bound by UTX with no alterations within 10 kb of the gene body.