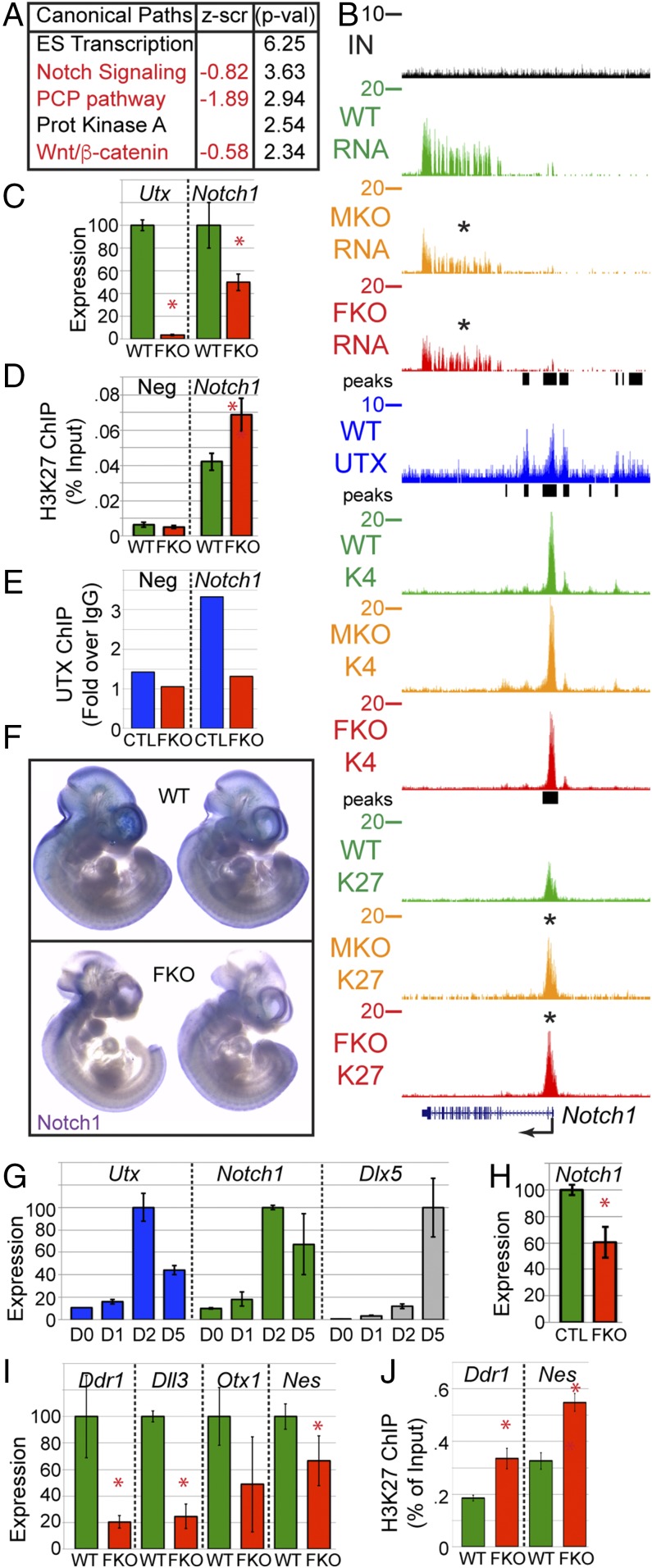
Fig. 5.
UTX regulates cranial NC targets. (A) Ingenuity Pathway Analysis of FKO-affected gene. Z score represents the overall alteration in FKO pathway expression. P = −log. (B) UCSC tracks of RNA-seq (RNA), H3K27me3 ChIP-seq (K27), H3K4me3 ChIP-seq (K4), and UTX ChIP-seq (UTX) at the Notch1 locus. (C) Quantitative RT-PCR of isolated primary NC (t test *P < 0.05, n ≥ 3). (D) H3K27me3 ChIP-qPCR. Neg is a control locus (Npm1) lacking H3K27me3 (t test *P < 0.03, n ≥ 3). (E) UTX ChIP-qPCR in O9-1 NC line treated with control (CTL) or Utx guide RNAs (FKO). Neg is a control locus (Slamf6). ChIP was normalized as percentage input and plotted as fold increase over IgG ChIP control. (F) Whole mount in situ hybridization for Notch1 in E10.5 WT or FKO embryos. (G) O9-1 maintained as NC stem cells (D0) or osteoprogenitor differentiated for indicated days. (H) Notch1 qRT-PCR at 2D of osteogenic differentiation with CRISPR for CTL or UTX FKO (t test *P < 0.03, n = 3). (I) Verification of RNA-seq targets by qRT-PCR (t test *P < 0.05, n ≥ 3). (J) Verification of H3K27me3 ChIP-seq targets by ChIP qPCR (t test *P < 0.02, n ≥ 3).