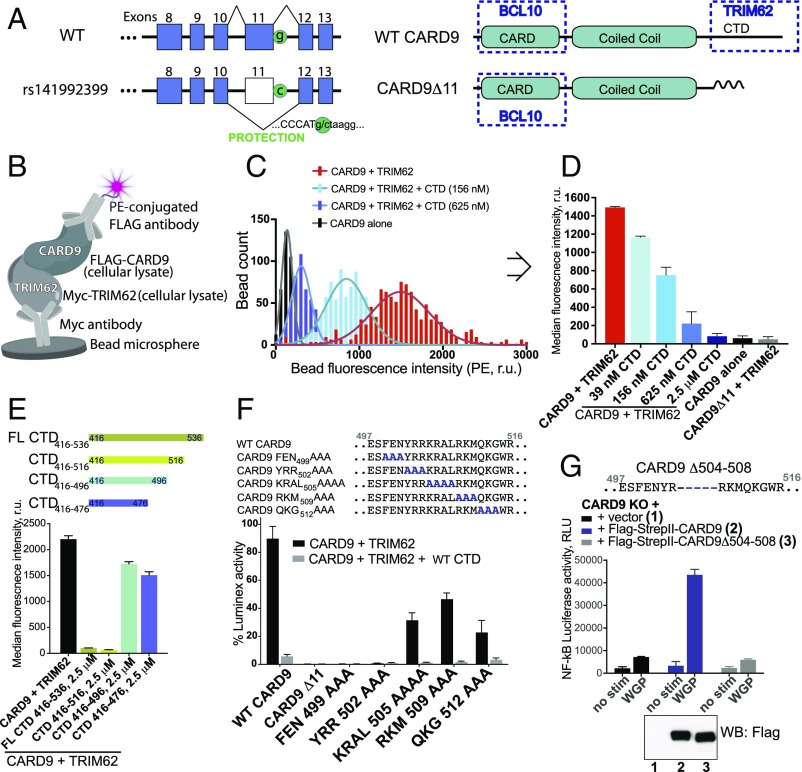
Fig. 1.
A highly sensitive bead-based ELISA reveals the key determinants of CARD9–TRIM62 interaction. (A) The CARD9Δ11 protective variant encodes a G-to-C substitution resulting in disruption of a splice site and exon 11 skipping. WT CARD9 consists of CARD domain (interacts with BCL10), coiled coil domain, and C-terminal domain (CTD) (binds TRIM62 during CARD9 activation, disrupted in the protective variant). (B) Bead-based assay designed to detect CARD9–TRIM62 PPI in vitro. (C) Histogram of fluorescence intensity of each bead for CARD9 and TRIM62 (red), CARD9, and TRIM62 in the presence of CTD (156 nM, light blue; 625 nM, dark blue), or CARD9 alone in the absence of TRIM62 (black). (D) Median fluorescence intensity is taken from integrated signals of all individual beads in a well and provides a robust measure of CARD9–TRIM62 PPI disruption, as evidenced by a dose-dependent CTD competition. CARD9Δ11 is unable to bind TRIM62. (E) CARD9–TRIM62 disruption by four truncated CTD constructs (CTD416–536, CTD416–516, CTD416–496, or CTD416–476). (F) Mutational mapping of key CARD9 residues responsible for TRIM62 binding. (G) Human variant CARD9Δ504–508 precisely corresponds to the key TRIM62-interacting residues and results in disrupted CARD9 signaling, as measured by Dectin-1–triggered NF-κB–driven luciferase activity in THP-1 cells. Western blot confirms equivalent protein expression. RLU, relative light units; r.u., relative units. Data in Fig. 1 are mean ± SD for at least triplicates.