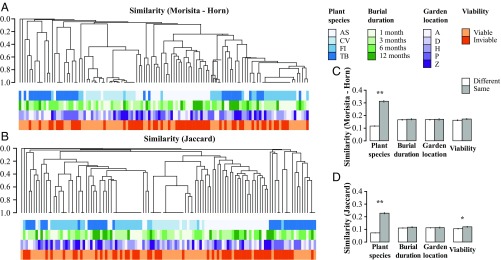
Fig. 1.
Hierarchical cluster analyses and pairwise similarities of seed-associated fungal communities reveal strong effects of plant species and seed viability on fungal community structure. Dendrograms represent Morisita–Horn (A) and Jaccard (B) similarities among communities of fungi. Tips of dendrograms represent fungal communities defined as the sequenced fungal strains isolated from seeds of a given plant species (AS, Annona spraguei; CV, Cochlospermum vitifolium; FI, Ficus insipida; and TB, Trema micrantha “black”), a specific burial duration (1, 3, 6, or 12 mo), a specific burial location (A, Armour; D, Drayton; H, 25 ha; P, Pearson; and Z, Zetek), and a specific viability class (viable or inviable seeds). Sequence data were obtained from 992 isolates from buried seeds (72%; mean ± SE, 110.2 ± 38.4 isolates per plant species; SI Appendix, Table S5). Together these represent 209 operational taxonomic units (OTU) based on 99% sequence similarity (Fisher’s alpha = 80.8). Overall, 107 OTU were represented by only one isolate (51.2%). Panels on the right summarize pairwise comparisons of fungal community similarity from the cluster diagrams based on abundance (C) and presence–absence data (D). Analysis of the entire dataset (nine plant species) also highlights the strong effect of plant species (SI Appendix, Fig. S6 and Tables S6 and S7). Error bars represent SE. **P < 0.001, *P < 0.05.