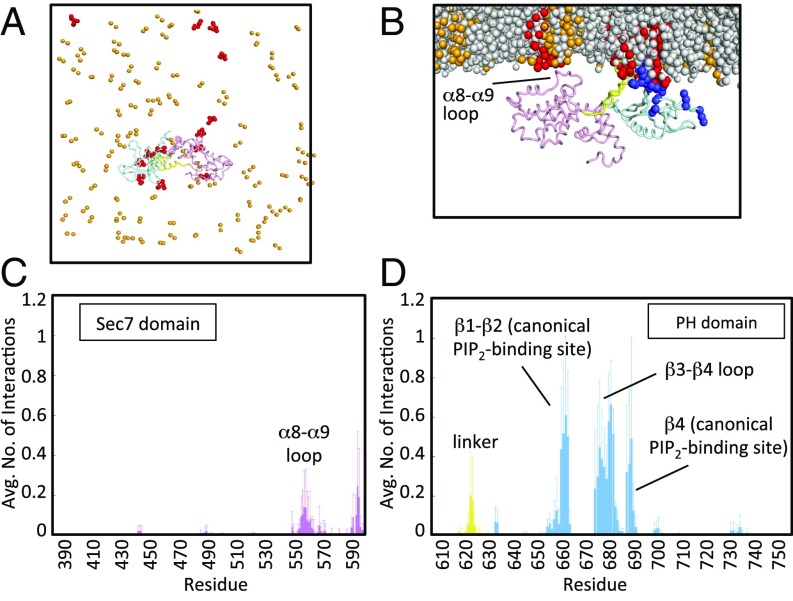
Fig. 2.
Coarse-grained MD simulations of Brag2 on a membrane showing multiple PIP2 lipid-binding sites. (A) Coarse-grained MD simulation of Brag2 on a membrane bilayer containing PIP2 lipids at the end of the simulation, for one run. Multiple PIP2s bind to the protein at various interaction sites at the end of the simulation. The PC lipids and the tails of the PS and PIP2 lipids are not shown for clarity. (B) Close-up of Brag2 at the end of the simulation run, for one run. The protein binds primarily to PIP2 molecules in the membrane at multiple binding sites. The view is rotated by 90° with respect to A. (C) Time-averaged number of interactions between Sec7 domain residues and PIP2 lipids. The Inset shows a close-up of the interactions of the residues in the α8-α9 loop. Additional interactions are also seen in the loop between the Sec7 domain and the linker (residues 580–599). (D) Time-averaged number of interactions between residues of the linker and the PH domain, and PIP2 lipids. See Fig. 1A and Fig. S1C for the localization of the structural elements bearing these residues. Positively charged residues (lysines and arginines) are in dark blue. PIP2 lipids are in red; PS lipids are in orange; PC lipids are in gray.