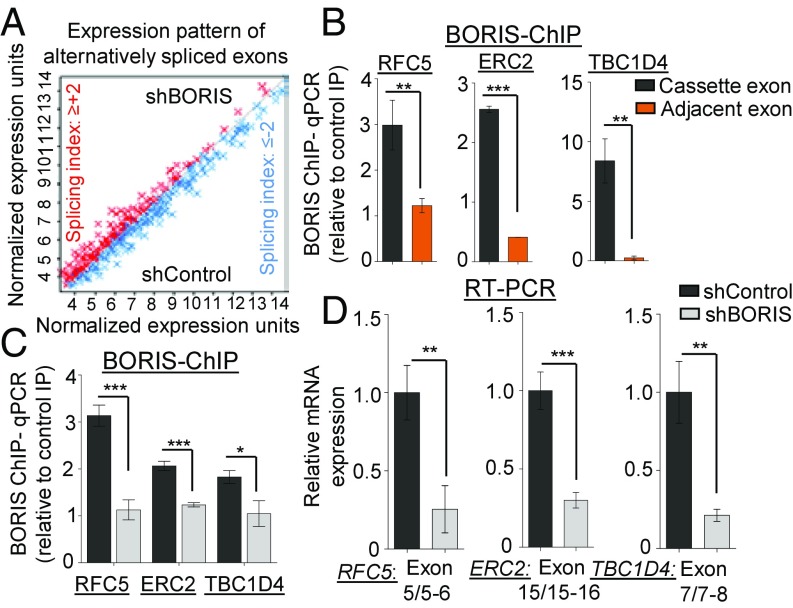
Fig. 8.
Global identification of BORIS-mediated alternative splicing. (A) Global differential expression pattern of alternatively spliced exons in shBORIS versus shControl transfected MCF7 cells [n = 2; excluded exons = 361 (splicing index ≤ −2), included exons = 221 (splicing index ≥ +2); and P < 0.05]. The graph represents normalized expression levels (signal space transformation–robust multiarray average algorithm) of differentially expressed exon probes. Red dots represent the exon inclusion; blue dots represent exon exclusion. (B) Validation of BBS in alternative exons by BORIS ChIP in MCF7 cells (n = 3). (C) BORIS ChIP-qPCR in MCF7 cells transfected with shBORIS and shControl cells to detect BORIS binding at alternative exons of genes from shBORIS versus shControl HTA 2.0 profile (n = 3). (D) Validation of BORIS-regulated alternative splicing events by qRT-PCR in shControl versus shBORIS transfected MCF7 cells (n = 3). Error bar shows mean values ± SD. As calculated using two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001.