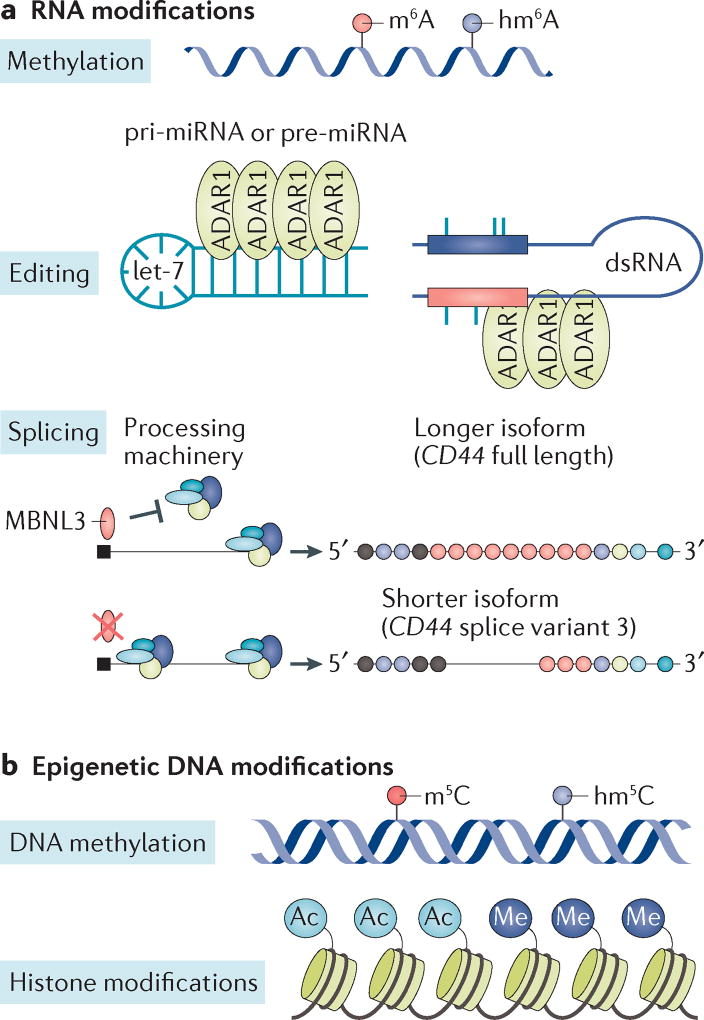
Figure 1. Epitranscriptome regulation contributes to cancer stem cell generation.
Normal haematopoietic stem cells (HSCs) accumulate genetic (DNA) and epitranscriptomic (RNA) changes that promote the emergence of pre-leukaemic clones that have gained survival and/or proliferative advantages. a | Deregulation of epitranscriptomic events include post-transcriptional RNA modifications such as RNA methylation (N6-methyladenosine (m6A) and N6-hydroxymethyladenosine (hm6A)) and adenosine-to-inosine (A-to-I) double-stranded RNA (dsRNA) editing (for example, adenosine deaminase acting on dsRNA 1 (ADAR1) editing of primary microRNA (pri-miRNA) and precursor miRNA (pre-miRNA) such as let-7). Alu element-containing dsRNA is the main target of A-to-I editing, which can result in changes to mRNA, secondary structure, stability and cellular localization. Splicing and the activity of RNA binding protein splicing factors such as muscleblind-like 3 (MBNL3) are additional RNA modifications. These events add additional layers of dynamic regulation that can all contribute to both cancer initiation and cancer progression. b | Epigenetic DNA modifications, such as the formation of 5-methylcytosine (m5C) and 5-hydroxymethylcytosine (hm5C), fulfil important roles in regulating cell differentiation and development. Genes with high levels of hm5C in their promoter regions are typically transcriptionally silent. This addition of methyl groups onto DNA will therefore alter the function of the DNA. For example, if a developmentally regulated gene has a high level of hm5C it may fail to induce differentiation and can potentially contribute to cancer initiation and progression. Histone modifications (methylation (Me) and acetylation (Ac)) are known to have important roles in cell differentiation and development with potential carcinogenic outcomes.